Project
Illumina Body Map 2
Navigation
Downloads
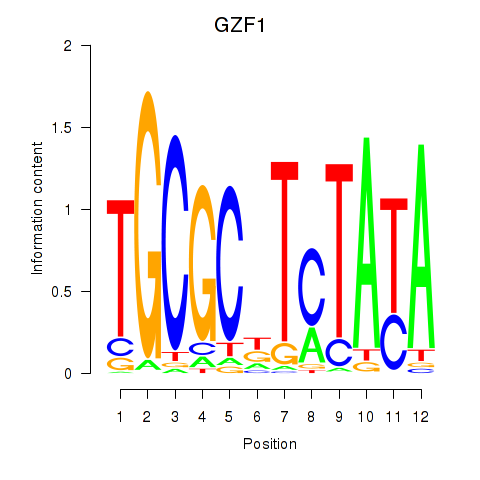
Results for GZF1
Z-value: 0.42
Transcription factors associated with GZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GZF1
|
ENSG00000125812.11 | GDNF inducible zinc finger protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GZF1 | hg19_v2_chr20_+_23342783_23342957 | -0.21 | 2.4e-01 | Click! |
Activity profile of GZF1 motif
Sorted Z-values of GZF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_27183263 | 1.93 |
ENST00000222726.3
|
HOXA5
|
homeobox A5 |
| chr12_+_54384370 | 1.36 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr3_-_87040233 | 1.19 |
ENST00000398399.2
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr3_-_87039662 | 1.07 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr3_+_30648066 | 0.75 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr3_-_87040259 | 0.73 |
ENST00000383698.3
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr3_+_30647994 | 0.72 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr3_-_79816965 | 0.69 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr14_+_22293618 | 0.65 |
ENST00000390432.2
|
TRAV10
|
T cell receptor alpha variable 10 |
| chr1_+_26606608 | 0.60 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr19_+_36157715 | 0.49 |
ENST00000379013.2
ENST00000222275.2 |
UPK1A
|
uroplakin 1A |
| chr14_-_52535712 | 0.47 |
ENST00000216286.5
ENST00000541773.1 |
NID2
|
nidogen 2 (osteonidogen) |
| chr11_+_101918153 | 0.47 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr5_+_92919043 | 0.46 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr5_+_170814803 | 0.44 |
ENST00000521672.1
ENST00000351986.6 ENST00000393820.2 ENST00000523622.1 |
NPM1
|
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
| chr1_+_9711781 | 0.42 |
ENST00000536656.1
ENST00000377346.4 |
PIK3CD
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr7_-_26904317 | 0.39 |
ENST00000345317.2
|
SKAP2
|
src kinase associated phosphoprotein 2 |
| chr1_-_201438282 | 0.37 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr3_-_170588163 | 0.36 |
ENST00000295830.8
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr14_-_92413353 | 0.35 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr19_-_59084647 | 0.35 |
ENST00000594234.1
ENST00000596039.1 |
MZF1
|
myeloid zinc finger 1 |
| chr8_+_23430157 | 0.35 |
ENST00000399967.3
|
FP15737
|
FP15737 |
| chr9_-_124976154 | 0.34 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr4_-_147443043 | 0.31 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr1_+_32757668 | 0.31 |
ENST00000373548.3
|
HDAC1
|
histone deacetylase 1 |
| chr9_-_124976185 | 0.30 |
ENST00000464484.2
|
LHX6
|
LIM homeobox 6 |
| chr14_-_92413727 | 0.28 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr19_-_59084922 | 0.28 |
ENST00000215057.2
ENST00000599369.1 |
MZF1
|
myeloid zinc finger 1 |
| chrX_+_153238220 | 0.27 |
ENST00000425274.1
|
TMEM187
|
transmembrane protein 187 |
| chr2_-_74776586 | 0.27 |
ENST00000420535.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr1_+_32757718 | 0.27 |
ENST00000428704.1
|
HDAC1
|
histone deacetylase 1 |
| chr12_-_76478386 | 0.27 |
ENST00000535020.2
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr19_-_14628645 | 0.26 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr12_-_76478446 | 0.26 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr12_-_76478417 | 0.26 |
ENST00000552342.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr6_-_42418999 | 0.25 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr4_-_147442817 | 0.25 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr16_+_67876180 | 0.25 |
ENST00000303596.1
|
THAP11
|
THAP domain containing 11 |
| chr9_-_14308004 | 0.24 |
ENST00000493697.1
|
NFIB
|
nuclear factor I/B |
| chr20_+_42086525 | 0.22 |
ENST00000244020.3
|
SRSF6
|
serine/arginine-rich splicing factor 6 |
| chr6_+_30525051 | 0.20 |
ENST00000376557.3
|
PRR3
|
proline rich 3 |
| chr19_-_36545128 | 0.19 |
ENST00000538849.1
|
THAP8
|
THAP domain containing 8 |
| chr6_+_30524663 | 0.18 |
ENST00000376560.3
|
PRR3
|
proline rich 3 |
| chr9_+_96338860 | 0.17 |
ENST00000375376.4
|
PHF2
|
PHD finger protein 2 |
| chr16_+_86612112 | 0.17 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr4_+_26344754 | 0.17 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_-_2461684 | 0.15 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr16_-_30582888 | 0.15 |
ENST00000563707.1
ENST00000567855.1 |
ZNF688
|
zinc finger protein 688 |
| chr8_+_112388852 | 0.13 |
ENST00000521904.1
ENST00000521753.1 |
RP11-1101K5.1
|
RP11-1101K5.1 |
| chr4_-_147442982 | 0.13 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr3_+_51705222 | 0.13 |
ENST00000457573.1
ENST00000341333.5 ENST00000412249.1 ENST00000425781.1 ENST00000415259.1 ENST00000395057.1 ENST00000416589.1 |
TEX264
|
testis expressed 264 |
| chr17_-_70588936 | 0.13 |
ENST00000453722.2
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr15_+_78370140 | 0.11 |
ENST00000409568.2
|
SH2D7
|
SH2 domain containing 7 |
| chr10_-_126849068 | 0.11 |
ENST00000494626.2
ENST00000337195.5 |
CTBP2
|
C-terminal binding protein 2 |
| chr12_+_102513950 | 0.09 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chrX_+_102883620 | 0.08 |
ENST00000372626.3
|
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr10_+_97759848 | 0.07 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr9_+_96338647 | 0.06 |
ENST00000359246.4
|
PHF2
|
PHD finger protein 2 |
| chr1_-_42384343 | 0.05 |
ENST00000372584.1
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr7_+_102004322 | 0.05 |
ENST00000496391.1
|
PRKRIP1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr19_-_3786253 | 0.05 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr10_+_89264625 | 0.05 |
ENST00000371996.4
ENST00000371994.4 |
MINPP1
|
multiple inositol-polyphosphate phosphatase 1 |
| chr13_+_98086445 | 0.05 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr19_-_3786354 | 0.04 |
ENST00000395040.2
ENST00000310132.6 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr6_+_7108210 | 0.04 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr16_-_70285797 | 0.04 |
ENST00000435634.1
|
EXOSC6
|
exosome component 6 |
| chr12_-_102513843 | 0.04 |
ENST00000551744.2
ENST00000552283.1 |
NUP37
|
nucleoporin 37kDa |
| chr22_+_50639408 | 0.04 |
ENST00000380903.2
|
SELO
|
Selenoprotein O |
| chr2_-_27603582 | 0.03 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr7_-_75115548 | 0.03 |
ENST00000453279.2
|
POM121C
|
POM121 transmembrane nucleoporin C |
| chr6_-_30524951 | 0.03 |
ENST00000376621.3
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr1_-_32801825 | 0.02 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr18_-_53303123 | 0.02 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr10_-_126849588 | 0.02 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr12_+_107168418 | 0.01 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr1_+_41174988 | 0.01 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr10_+_14880287 | 0.01 |
ENST00000437161.2
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr12_-_120662499 | 0.00 |
ENST00000552550.1
|
PXN
|
paxillin |
| chr19_-_10426663 | 0.00 |
ENST00000541276.1
ENST00000393708.3 ENST00000494368.1 |
FDX1L
|
ferredoxin 1-like |
| chr17_-_29648761 | 0.00 |
ENST00000247270.3
ENST00000462804.2 |
EVI2A
|
ecotropic viral integration site 2A |
Network of associatons between targets according to the STRING database.
First level regulatory network of GZF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.5 | 1.5 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.7 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.1 | 0.6 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.4 | GO:0060374 | positive regulation of neutrophil apoptotic process(GO:0033031) mast cell differentiation(GO:0060374) |
| 0.1 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.6 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:0060699 | regulation of endoribonuclease activity(GO:0060699) |
| 0.0 | 0.2 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.2 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.5 | GO:0071711 | basement membrane organization(GO:0071711) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 3.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |