Project
Illumina Body Map 2
Navigation
Downloads
Results for HDX
Z-value: 0.47
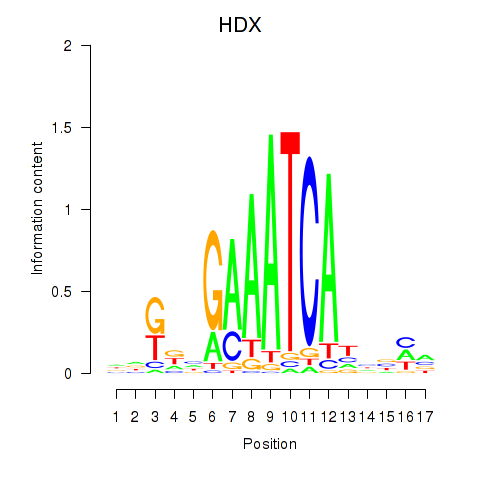
Transcription factors associated with HDX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HDX
|
ENSG00000165259.9 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HDX | hg19_v2_chrX_-_83757399_83757487 | -0.14 | 4.6e-01 | Click! |
Activity profile of HDX motif
Sorted Z-values of HDX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_159684371 | 1.79 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr5_-_41213607 | 1.77 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr12_-_103344615 | 1.70 |
ENST00000546844.1
|
PAH
|
phenylalanine hydroxylase |
| chr15_-_54025300 | 1.44 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr3_-_165555200 | 1.29 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr5_-_135290651 | 1.27 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr5_-_35230649 | 0.99 |
ENST00000382002.5
|
PRLR
|
prolactin receptor |
| chr5_-_35230771 | 0.94 |
ENST00000342362.5
|
PRLR
|
prolactin receptor |
| chr5_-_135290705 | 0.90 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr7_-_121944491 | 0.85 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr5_-_35230434 | 0.82 |
ENST00000504500.1
|
PRLR
|
prolactin receptor |
| chr11_-_66675371 | 0.79 |
ENST00000393955.2
|
PC
|
pyruvate carboxylase |
| chr2_+_128403439 | 0.75 |
ENST00000544369.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr2_+_128403720 | 0.72 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr1_+_21880560 | 0.63 |
ENST00000425315.2
|
ALPL
|
alkaline phosphatase, liver/bone/kidney |
| chr12_+_123849462 | 0.61 |
ENST00000543072.1
|
hsa-mir-8072
|
hsa-mir-8072 |
| chr6_-_116381918 | 0.56 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr10_-_21186144 | 0.49 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr5_-_35230569 | 0.48 |
ENST00000515839.1
|
PRLR
|
prolactin receptor |
| chr16_+_1877204 | 0.44 |
ENST00000427358.2
|
FAHD1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr10_-_14372870 | 0.44 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr2_+_113735575 | 0.42 |
ENST00000376489.2
ENST00000259205.4 |
IL36G
|
interleukin 36, gamma |
| chr19_-_58485895 | 0.40 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr9_+_6215799 | 0.34 |
ENST00000417746.2
ENST00000456383.2 |
IL33
|
interleukin 33 |
| chr1_-_209825674 | 0.32 |
ENST00000367030.3
ENST00000356082.4 |
LAMB3
|
laminin, beta 3 |
| chr4_+_48807155 | 0.29 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr2_-_40657397 | 0.29 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr12_-_39734783 | 0.27 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr2_+_102758271 | 0.24 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr8_+_145065705 | 0.24 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr13_+_24144796 | 0.21 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr1_+_84630367 | 0.19 |
ENST00000370680.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_+_84630352 | 0.17 |
ENST00000450730.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_+_102758210 | 0.17 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr12_+_101869096 | 0.17 |
ENST00000551346.1
|
SPIC
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr19_+_36602104 | 0.13 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr3_+_107241882 | 0.13 |
ENST00000416476.2
|
BBX
|
bobby sox homolog (Drosophila) |
| chr12_-_15815626 | 0.12 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr1_+_47489240 | 0.10 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr6_+_98264571 | 0.10 |
ENST00000607823.1
|
RP11-436D23.1
|
RP11-436D23.1 |
| chr18_+_29027696 | 0.10 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr5_+_140165876 | 0.09 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr5_+_140625147 | 0.07 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr1_+_84629976 | 0.06 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr13_+_24144509 | 0.06 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chrX_+_27608490 | 0.06 |
ENST00000451261.2
|
DCAF8L2
|
DDB1 and CUL4 associated factor 8-like 2 |
| chr12_+_10460417 | 0.05 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr12_-_31743901 | 0.04 |
ENST00000354285.4
|
DENND5B
|
DENN/MADD domain containing 5B |
| chrX_+_27608517 | 0.04 |
ENST00000431122.2
|
DCAF8L2
|
DDB1 and CUL4 associated factor 8-like 2 |
| chr14_+_79745682 | 0.03 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr18_-_53804580 | 0.03 |
ENST00000590484.1
ENST00000589293.1 ENST00000587904.1 ENST00000591974.1 |
RP11-456O19.4
|
RP11-456O19.4 |
| chr14_+_79745746 | 0.01 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr22_+_40440804 | 0.01 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
Network of associatons between targets according to the STRING database.
First level regulatory network of HDX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.4 | 3.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.4 | 1.8 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.2 | 0.6 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.2 | 0.8 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 1.8 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.1 | 1.7 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 1.5 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.3 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 1.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.4 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.6 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 3.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.5 | 3.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 1.7 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.4 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.8 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 0.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 1.5 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.5 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |