Project
Illumina Body Map 2
Navigation
Downloads
Results for HINFP
Z-value: 1.03
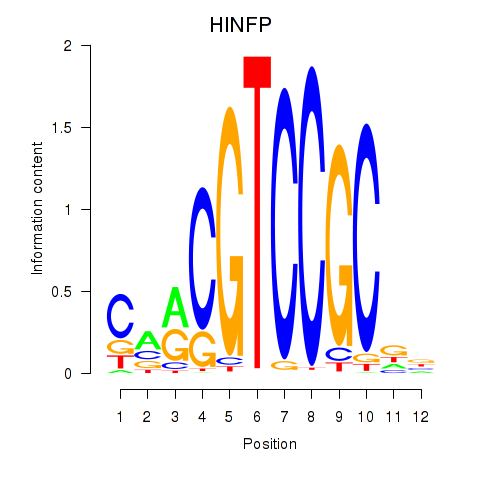
Transcription factors associated with HINFP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HINFP
|
ENSG00000172273.8 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HINFP | hg19_v2_chr11_+_118992269_118992334 | -0.12 | 5.0e-01 | Click! |
Activity profile of HINFP motif
Sorted Z-values of HINFP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_54962704 | 1.71 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr3_-_38691119 | 1.60 |
ENST00000333535.4
ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit |
| chr16_-_54962415 | 1.55 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr16_-_54962625 | 1.50 |
ENST00000559432.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr1_-_40157345 | 1.34 |
ENST00000372844.3
|
HPCAL4
|
hippocalcin like 4 |
| chr22_+_18593446 | 1.27 |
ENST00000316027.6
|
TUBA8
|
tubulin, alpha 8 |
| chr18_-_44336998 | 1.26 |
ENST00000315087.7
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr22_+_18593507 | 1.18 |
ENST00000330423.3
|
TUBA8
|
tubulin, alpha 8 |
| chr17_+_38024417 | 1.16 |
ENST00000348931.4
ENST00000583811.1 ENST00000584588.1 ENST00000377940.3 |
ZPBP2
|
zona pellucida binding protein 2 |
| chr1_-_217262969 | 1.12 |
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr22_+_18593097 | 1.07 |
ENST00000426208.1
|
TUBA8
|
tubulin, alpha 8 |
| chr19_+_36545833 | 1.07 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr5_-_127873496 | 1.06 |
ENST00000508989.1
|
FBN2
|
fibrillin 2 |
| chr20_-_30458432 | 1.04 |
ENST00000375966.4
ENST00000278979.3 |
DUSP15
|
dual specificity phosphatase 15 |
| chr1_-_217262933 | 1.03 |
ENST00000359162.2
|
ESRRG
|
estrogen-related receptor gamma |
| chr16_-_54963026 | 1.03 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr20_-_30458019 | 1.01 |
ENST00000486996.1
ENST00000398084.2 |
DUSP15
|
dual specificity phosphatase 15 |
| chr17_+_30593195 | 0.95 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr2_+_120302041 | 0.94 |
ENST00000442513.3
ENST00000413369.3 |
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr5_-_88180342 | 0.94 |
ENST00000502983.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr20_-_30458491 | 0.93 |
ENST00000339738.5
|
DUSP15
|
dual specificity phosphatase 15 |
| chr5_-_41510656 | 0.92 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr16_+_6069586 | 0.91 |
ENST00000547372.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr5_+_36606700 | 0.90 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr20_-_30458354 | 0.89 |
ENST00000428829.1
|
DUSP15
|
dual specificity phosphatase 15 |
| chr6_+_159291090 | 0.89 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr5_-_41510725 | 0.89 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr6_+_146350622 | 0.88 |
ENST00000507907.1
|
GRM1
|
glutamate receptor, metabotropic 1 |
| chr5_+_36606992 | 0.85 |
ENST00000505202.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr15_-_35047166 | 0.85 |
ENST00000290374.4
|
GJD2
|
gap junction protein, delta 2, 36kDa |
| chr4_+_89514402 | 0.84 |
ENST00000426683.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr13_+_111805980 | 0.82 |
ENST00000491775.1
ENST00000466143.1 ENST00000544132.1 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr14_+_90528109 | 0.79 |
ENST00000282146.4
|
KCNK13
|
potassium channel, subfamily K, member 13 |
| chr6_+_159290917 | 0.79 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr4_+_89514516 | 0.78 |
ENST00000452979.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr17_-_45056606 | 0.77 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr5_-_132073111 | 0.76 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr2_+_120301997 | 0.76 |
ENST00000602047.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr10_-_128994422 | 0.73 |
ENST00000522781.1
|
FAM196A
|
family with sequence similarity 196, member A |
| chr9_+_34458771 | 0.70 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr10_-_62761188 | 0.69 |
ENST00000357917.4
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chrX_-_25034065 | 0.68 |
ENST00000379044.4
|
ARX
|
aristaless related homeobox |
| chr14_-_101034811 | 0.67 |
ENST00000553553.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr2_-_233352531 | 0.66 |
ENST00000304546.1
|
ECEL1
|
endothelin converting enzyme-like 1 |
| chr11_+_64879317 | 0.65 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr15_+_43803143 | 0.64 |
ENST00000382031.1
|
MAP1A
|
microtubule-associated protein 1A |
| chr16_+_81069433 | 0.64 |
ENST00000299575.4
|
ATMIN
|
ATM interactor |
| chr3_+_50654550 | 0.64 |
ENST00000430409.1
ENST00000357955.2 |
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr2_-_232791038 | 0.63 |
ENST00000295440.2
ENST00000409852.1 |
NPPC
|
natriuretic peptide C |
| chr19_+_18077881 | 0.62 |
ENST00000609922.1
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr4_+_91048706 | 0.62 |
ENST00000509176.1
|
CCSER1
|
coiled-coil serine-rich protein 1 |
| chr4_-_6202247 | 0.62 |
ENST00000409021.3
ENST00000409371.3 |
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr16_-_28074822 | 0.61 |
ENST00000395724.3
ENST00000380898.2 ENST00000447459.2 |
GSG1L
|
GSG1-like |
| chr2_+_157292859 | 0.60 |
ENST00000438166.2
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr1_-_197115818 | 0.59 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr5_+_61602236 | 0.59 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr3_+_50654821 | 0.59 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr4_-_6202291 | 0.59 |
ENST00000282924.5
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr6_+_56819895 | 0.58 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr5_+_98264867 | 0.58 |
ENST00000513175.1
|
CTD-2007H13.3
|
CTD-2007H13.3 |
| chr17_+_17206635 | 0.57 |
ENST00000389022.4
|
NT5M
|
5',3'-nucleotidase, mitochondrial |
| chr7_-_29234802 | 0.57 |
ENST00000449801.1
ENST00000409850.1 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr1_+_214454492 | 0.57 |
ENST00000366957.5
ENST00000415093.2 |
SMYD2
|
SET and MYND domain containing 2 |
| chr2_+_30670209 | 0.57 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr5_-_132073210 | 0.57 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr2_-_170219037 | 0.57 |
ENST00000443831.1
|
LRP2
|
low density lipoprotein receptor-related protein 2 |
| chr9_+_130965677 | 0.57 |
ENST00000393594.3
ENST00000486160.1 |
DNM1
|
dynamin 1 |
| chr9_+_130965651 | 0.56 |
ENST00000475805.1
ENST00000341179.7 ENST00000372923.3 |
DNM1
|
dynamin 1 |
| chr13_-_48877795 | 0.50 |
ENST00000436963.1
ENST00000433480.2 |
LINC00441
|
long intergenic non-protein coding RNA 441 |
| chr22_-_22221658 | 0.50 |
ENST00000544786.1
|
MAPK1
|
mitogen-activated protein kinase 1 |
| chr17_-_42276574 | 0.50 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr2_+_30670127 | 0.49 |
ENST00000540623.1
ENST00000476038.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr17_+_11501748 | 0.49 |
ENST00000262442.4
ENST00000579828.1 |
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr9_+_37650945 | 0.48 |
ENST00000377765.3
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr5_+_61601965 | 0.48 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chr5_+_142149955 | 0.48 |
ENST00000378004.3
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr22_-_22221900 | 0.48 |
ENST00000215832.6
ENST00000398822.3 |
MAPK1
|
mitogen-activated protein kinase 1 |
| chr8_-_66754172 | 0.48 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr2_-_170219079 | 0.48 |
ENST00000263816.3
|
LRP2
|
low density lipoprotein receptor-related protein 2 |
| chr1_+_6845384 | 0.47 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr5_+_143584814 | 0.47 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr14_-_81902791 | 0.47 |
ENST00000557055.1
|
STON2
|
stonin 2 |
| chr7_+_29234375 | 0.46 |
ENST00000409350.1
ENST00000495789.2 ENST00000539389.1 |
CHN2
|
chimerin 2 |
| chr11_-_132813627 | 0.46 |
ENST00000374778.4
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr2_+_120302055 | 0.45 |
ENST00000598644.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr16_+_84002234 | 0.44 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr5_+_61602055 | 0.44 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr11_-_132813566 | 0.44 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr4_-_156298028 | 0.43 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr8_-_9760839 | 0.43 |
ENST00000519461.1
ENST00000517675.1 |
LINC00599
|
long intergenic non-protein coding RNA 599 |
| chr18_+_54814288 | 0.43 |
ENST00000585477.1
|
BOD1L2
|
biorientation of chromosomes in cell division 1-like 2 |
| chr19_-_14016877 | 0.41 |
ENST00000454313.1
ENST00000591586.1 ENST00000346736.2 |
C19orf57
|
chromosome 19 open reading frame 57 |
| chr14_+_64971438 | 0.40 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr20_+_3451732 | 0.40 |
ENST00000446916.2
|
ATRN
|
attractin |
| chr5_+_142149932 | 0.39 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chrX_+_21392553 | 0.39 |
ENST00000279451.4
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr3_+_94657016 | 0.39 |
ENST00000462219.1
|
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr6_+_24495185 | 0.39 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr20_-_32031680 | 0.38 |
ENST00000217381.2
|
SNTA1
|
syntrophin, alpha 1 |
| chr8_-_142318398 | 0.38 |
ENST00000520137.1
|
SLC45A4
|
solute carrier family 45, member 4 |
| chr1_-_51796226 | 0.38 |
ENST00000451380.1
ENST00000371747.3 ENST00000439482.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr1_-_2706236 | 0.38 |
ENST00000401095.3
ENST00000574621.2 |
TTC34
|
tetratricopeptide repeat domain 34 |
| chr14_-_81902516 | 0.38 |
ENST00000554710.1
|
STON2
|
stonin 2 |
| chr2_+_30670077 | 0.37 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr17_-_1419941 | 0.37 |
ENST00000498390.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr19_+_36545781 | 0.37 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chrX_-_134232630 | 0.37 |
ENST00000535837.1
ENST00000433425.2 |
LINC00087
|
long intergenic non-protein coding RNA 87 |
| chr3_+_63897605 | 0.37 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr2_+_204193149 | 0.37 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chrX_+_21392529 | 0.36 |
ENST00000425654.2
ENST00000543067.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr1_+_61547405 | 0.36 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr17_+_11501816 | 0.36 |
ENST00000454412.2
|
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr8_+_32406137 | 0.36 |
ENST00000521670.1
|
NRG1
|
neuregulin 1 |
| chr13_+_25875785 | 0.35 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr2_+_204193129 | 0.34 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr16_+_4526341 | 0.34 |
ENST00000458134.3
ENST00000219700.6 ENST00000575120.1 ENST00000572812.1 ENST00000574466.1 ENST00000576827.1 ENST00000570445.1 |
HMOX2
|
heme oxygenase (decycling) 2 |
| chr9_-_115095229 | 0.33 |
ENST00000210227.4
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr9_-_139922631 | 0.33 |
ENST00000341511.6
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr12_-_90103077 | 0.33 |
ENST00000551310.1
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr13_-_22178284 | 0.33 |
ENST00000468222.2
ENST00000382374.4 |
MICU2
|
mitochondrial calcium uptake 2 |
| chr2_-_73053126 | 0.33 |
ENST00000272427.6
ENST00000410104.1 |
EXOC6B
|
exocyst complex component 6B |
| chr9_+_129622904 | 0.33 |
ENST00000319119.4
|
ZBTB34
|
zinc finger and BTB domain containing 34 |
| chr13_-_48612067 | 0.32 |
ENST00000543413.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr9_+_77112244 | 0.32 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr3_-_113415441 | 0.32 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr1_+_109102652 | 0.32 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr13_-_36705425 | 0.31 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr17_+_46125685 | 0.31 |
ENST00000579889.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr2_-_37384175 | 0.31 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr11_-_73309112 | 0.31 |
ENST00000450446.2
|
FAM168A
|
family with sequence similarity 168, member A |
| chr11_-_46867780 | 0.31 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chrX_-_38186811 | 0.30 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr2_-_68479614 | 0.30 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr11_+_1411503 | 0.29 |
ENST00000526678.1
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr6_+_56820018 | 0.29 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chr10_+_75532028 | 0.28 |
ENST00000372841.3
ENST00000394790.1 |
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
| chr17_+_29158962 | 0.28 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chrX_+_71996972 | 0.28 |
ENST00000334036.5
|
DMRTC1B
|
DMRT-like family C1B |
| chr7_-_134143841 | 0.28 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr22_-_20255212 | 0.28 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr9_-_139922726 | 0.28 |
ENST00000265662.5
ENST00000371605.3 |
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr11_+_119076745 | 0.28 |
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr11_-_6440624 | 0.27 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr20_+_3451650 | 0.27 |
ENST00000262919.5
|
ATRN
|
attractin |
| chrX_+_17393543 | 0.27 |
ENST00000380060.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr17_+_43972010 | 0.27 |
ENST00000334239.8
ENST00000446361.3 |
MAPT
|
microtubule-associated protein tau |
| chr9_-_98279241 | 0.27 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr11_-_6440283 | 0.26 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr17_-_1419878 | 0.26 |
ENST00000449479.1
ENST00000477910.1 ENST00000542125.1 ENST00000575172.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr5_-_136834982 | 0.26 |
ENST00000510689.1
ENST00000394945.1 |
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr19_-_35323762 | 0.26 |
ENST00000590963.1
|
CTC-523E23.4
|
CTC-523E23.4 |
| chr15_+_91643442 | 0.26 |
ENST00000394232.1
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr15_+_91643173 | 0.25 |
ENST00000545111.2
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr20_-_35492048 | 0.25 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr10_-_118502070 | 0.25 |
ENST00000369209.3
|
HSPA12A
|
heat shock 70kDa protein 12A |
| chr3_-_13009168 | 0.24 |
ENST00000273221.4
|
IQSEC1
|
IQ motif and Sec7 domain 1 |
| chr2_+_28974489 | 0.24 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr19_-_39523165 | 0.24 |
ENST00000509137.2
ENST00000292853.4 |
FBXO27
|
F-box protein 27 |
| chr13_-_99738867 | 0.23 |
ENST00000427887.1
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr2_-_33824336 | 0.23 |
ENST00000431950.1
ENST00000403368.1 ENST00000441530.2 |
FAM98A
|
family with sequence similarity 98, member A |
| chr3_-_156272924 | 0.23 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr12_+_56137064 | 0.22 |
ENST00000257868.5
ENST00000546799.1 |
GDF11
|
growth differentiation factor 11 |
| chr16_+_2510081 | 0.22 |
ENST00000361837.4
ENST00000569496.1 ENST00000567489.1 ENST00000563531.1 ENST00000483320.1 |
C16orf59
|
chromosome 16 open reading frame 59 |
| chr13_-_77460525 | 0.22 |
ENST00000377474.2
ENST00000317765.2 |
KCTD12
|
potassium channel tetramerization domain containing 12 |
| chr3_-_52312636 | 0.22 |
ENST00000296490.3
|
WDR82
|
WD repeat domain 82 |
| chr10_+_94608218 | 0.21 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr6_-_139695757 | 0.21 |
ENST00000367651.2
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr10_+_94352956 | 0.21 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr12_-_90102594 | 0.21 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chrX_-_38186775 | 0.21 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr15_-_83953466 | 0.21 |
ENST00000345382.2
|
BNC1
|
basonuclin 1 |
| chr17_+_7341586 | 0.20 |
ENST00000575235.1
|
FGF11
|
fibroblast growth factor 11 |
| chr3_-_196295385 | 0.20 |
ENST00000425888.1
|
WDR53
|
WD repeat domain 53 |
| chr18_-_268019 | 0.20 |
ENST00000261600.6
|
THOC1
|
THO complex 1 |
| chr1_-_36915880 | 0.20 |
ENST00000445843.3
|
OSCP1
|
organic solute carrier partner 1 |
| chr2_-_54087066 | 0.19 |
ENST00000394705.2
ENST00000352846.3 ENST00000406625.2 |
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr17_-_1419914 | 0.19 |
ENST00000397335.3
ENST00000574561.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr9_-_123555655 | 0.19 |
ENST00000340778.5
ENST00000453291.1 ENST00000608872.1 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr11_+_65819802 | 0.19 |
ENST00000528302.1
ENST00000322535.6 ENST00000524627.1 ENST00000533595.1 ENST00000530322.1 |
SF3B2
|
splicing factor 3b, subunit 2, 145kDa |
| chr12_+_69633407 | 0.19 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr12_-_99288536 | 0.19 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_+_196295482 | 0.19 |
ENST00000440469.1
ENST00000311630.6 |
FBXO45
|
F-box protein 45 |
| chr2_+_171785824 | 0.19 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr5_+_72143988 | 0.19 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr3_-_52719810 | 0.19 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr2_-_128785619 | 0.18 |
ENST00000450957.1
|
SAP130
|
Sin3A-associated protein, 130kDa |
| chr9_-_122131696 | 0.18 |
ENST00000373964.2
ENST00000265922.3 |
BRINP1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr4_-_186732892 | 0.18 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_+_49761147 | 0.18 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr17_-_66453562 | 0.18 |
ENST00000262139.5
ENST00000546360.1 |
WIPI1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr9_+_139921916 | 0.18 |
ENST00000314330.2
|
C9orf139
|
chromosome 9 open reading frame 139 |
| chr12_-_117175819 | 0.18 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr12_+_53773944 | 0.18 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr13_+_111806055 | 0.18 |
ENST00000218789.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr8_-_144241432 | 0.18 |
ENST00000430474.2
|
LY6H
|
lymphocyte antigen 6 complex, locus H |
| chr17_-_42277203 | 0.18 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr3_-_196295437 | 0.17 |
ENST00000429115.1
|
WDR53
|
WD repeat domain 53 |
| chr9_+_100174232 | 0.17 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr11_-_73309228 | 0.17 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chrX_-_53711064 | 0.17 |
ENST00000342160.3
ENST00000446750.1 |
HUWE1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
Network of associatons between targets according to the STRING database.
First level regulatory network of HINFP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 1.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 1.6 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.2 | 0.6 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.5 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.2 | 1.1 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.2 | 2.3 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.2 | 0.5 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 1.4 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 0.9 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.5 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 0.6 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.4 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 1.7 | GO:0072386 | plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.4 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.3 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.6 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.9 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.6 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.7 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.3 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.1 | 1.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 3.9 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.1 | 0.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 1.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.5 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.4 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 0.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 1.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.2 | GO:0001928 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.3 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.7 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.3 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.2 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 0.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.4 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.4 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 1.4 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.3 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.6 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 1.0 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 2.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 1.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0090240 | follicle-stimulating hormone signaling pathway(GO:0042699) positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 1.0 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.5 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.3 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.8 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.7 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.9 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.7 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 0.9 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 1.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.5 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 1.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 1.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.6 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 1.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 1.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.3 | 2.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 0.6 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.2 | 1.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.6 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 1.0 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 1.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.5 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 1.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.4 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.3 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.4 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.9 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.3 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 2.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 2.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 4.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 1.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |