Project
Illumina Body Map 2
Navigation
Downloads
Results for HMBOX1
Z-value: 0.90
Transcription factors associated with HMBOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMBOX1
|
ENSG00000147421.13 | homeobox containing 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMBOX1 | hg19_v2_chr8_+_28747884_28747938 | -0.29 | 1.0e-01 | Click! |
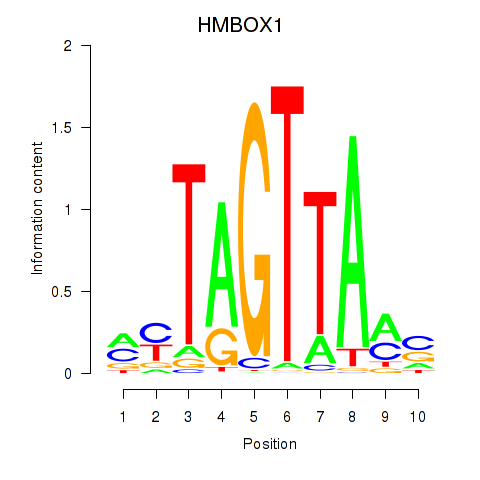
Activity profile of HMBOX1 motif
Sorted Z-values of HMBOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_71610985 | 5.19 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr20_-_7238861 | 4.39 |
ENST00000428954.1
|
RP11-19D2.1
|
RP11-19D2.1 |
| chr20_+_42187682 | 4.37 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr20_+_42187608 | 3.73 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr6_-_107235331 | 3.56 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr6_-_52668605 | 2.97 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr6_-_107235287 | 2.77 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr7_-_120498357 | 2.75 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr10_+_115312766 | 2.50 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr8_-_63951730 | 2.41 |
ENST00000260118.6
|
GGH
|
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) |
| chr10_-_54531406 | 2.34 |
ENST00000373968.3
|
MBL2
|
mannose-binding lectin (protein C) 2, soluble |
| chr4_+_187187098 | 2.24 |
ENST00000403665.2
ENST00000264692.4 |
F11
|
coagulation factor XI |
| chr6_-_52628271 | 2.13 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr9_-_104145795 | 2.13 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr4_-_110723134 | 2.07 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr4_-_110723194 | 1.99 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr8_+_30244580 | 1.86 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr4_+_156824840 | 1.76 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr4_-_69434245 | 1.76 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr10_-_5046042 | 1.73 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chrM_+_5824 | 1.65 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr2_+_182850551 | 1.63 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr11_+_19798964 | 1.61 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr4_+_187187337 | 1.56 |
ENST00000492972.2
|
F11
|
coagulation factor XI |
| chrX_+_66764375 | 1.54 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr1_+_196743912 | 1.49 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr4_+_74347400 | 1.48 |
ENST00000226355.3
|
AFM
|
afamin |
| chr1_+_196743943 | 1.44 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr3_+_119501557 | 1.41 |
ENST00000337940.4
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr12_-_33049690 | 1.34 |
ENST00000070846.6
ENST00000340811.4 |
PKP2
|
plakophilin 2 |
| chr20_+_61584026 | 1.32 |
ENST00000370351.4
ENST00000370349.3 |
SLC17A9
|
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
| chr9_+_101984577 | 1.32 |
ENST00000223641.4
|
SEC61B
|
Sec61 beta subunit |
| chr2_-_133429091 | 1.28 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr5_+_92919043 | 1.26 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr12_-_22063787 | 1.23 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr4_-_152149033 | 1.23 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr10_+_89419370 | 1.21 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr15_+_69857515 | 1.20 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr4_-_186877481 | 1.19 |
ENST00000444781.1
ENST00000432655.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_-_7553852 | 1.15 |
ENST00000593547.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chrX_-_10851762 | 1.14 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr4_-_186877502 | 1.10 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_112408661 | 1.08 |
ENST00000368662.5
|
TUBE1
|
tubulin, epsilon 1 |
| chr1_+_175036966 | 1.08 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr12_-_77272765 | 1.06 |
ENST00000547435.1
ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2
|
cysteine and glycine-rich protein 2 |
| chr19_-_7553889 | 1.05 |
ENST00000221480.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr9_+_139377947 | 1.04 |
ENST00000354376.1
|
C9orf163
|
chromosome 9 open reading frame 163 |
| chr19_+_50084561 | 1.03 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr11_+_22694123 | 1.01 |
ENST00000534801.1
|
GAS2
|
growth arrest-specific 2 |
| chr18_-_5197501 | 0.99 |
ENST00000580650.1
|
C18orf42
|
chromosome 18 open reading frame 42 |
| chr2_-_197226875 | 0.98 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr3_-_149095652 | 0.98 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr4_-_140477928 | 0.97 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr17_-_38083092 | 0.94 |
ENST00000394169.1
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae) |
| chr11_-_59950519 | 0.93 |
ENST00000528851.1
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chrX_+_16964985 | 0.89 |
ENST00000303843.7
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr7_+_117251671 | 0.89 |
ENST00000468795.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr11_-_59950622 | 0.84 |
ENST00000323961.3
ENST00000412309.2 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr11_-_59950486 | 0.84 |
ENST00000426738.2
ENST00000533023.1 ENST00000420732.2 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr6_+_123038689 | 0.79 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr12_+_94071341 | 0.79 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr6_+_112408768 | 0.79 |
ENST00000368656.2
ENST00000604268.1 |
FAM229B
|
family with sequence similarity 229, member B |
| chrX_-_15683147 | 0.78 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr6_+_138699042 | 0.77 |
ENST00000573100.1
|
RP3-422G23.4
|
Uncharacterized protein |
| chr17_-_56494908 | 0.75 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr17_-_56494882 | 0.75 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr16_-_3422283 | 0.72 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr2_-_3521518 | 0.71 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chrX_+_114874727 | 0.71 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr5_-_133968459 | 0.69 |
ENST00000505758.1
ENST00000439578.1 ENST00000502286.1 |
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr14_+_64854958 | 0.67 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr11_+_34643600 | 0.66 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr10_+_75936444 | 0.66 |
ENST00000372734.3
ENST00000541550.1 |
ADK
|
adenosine kinase |
| chr1_-_198906528 | 0.66 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr6_-_52710893 | 0.66 |
ENST00000284562.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr2_+_159825143 | 0.65 |
ENST00000454300.1
ENST00000263635.6 |
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr1_-_201346761 | 0.65 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr6_+_138725343 | 0.64 |
ENST00000607197.1
ENST00000367697.3 |
HEBP2
|
heme binding protein 2 |
| chr6_+_30585486 | 0.64 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr4_-_114900831 | 0.62 |
ENST00000315366.7
|
ARSJ
|
arylsulfatase family, member J |
| chr17_+_41177220 | 0.62 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr14_+_75536280 | 0.62 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr12_-_56236734 | 0.61 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chr1_+_170115142 | 0.59 |
ENST00000439373.2
|
METTL11B
|
methyltransferase like 11B |
| chr11_-_57194948 | 0.59 |
ENST00000533235.1
ENST00000526621.1 ENST00000352187.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chrX_-_99987088 | 0.59 |
ENST00000372981.1
ENST00000263033.5 |
SYTL4
|
synaptotagmin-like 4 |
| chr14_+_75536335 | 0.57 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr9_-_101984184 | 0.57 |
ENST00000476832.1
|
ALG2
|
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chr7_+_134576151 | 0.57 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr17_-_26903900 | 0.56 |
ENST00000395319.3
ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC
|
aldolase C, fructose-bisphosphate |
| chr11_+_17281900 | 0.55 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chr11_+_124789146 | 0.55 |
ENST00000408930.5
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1 |
| chr4_-_140477910 | 0.54 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr11_+_100862811 | 0.54 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr11_-_111649074 | 0.52 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr1_-_9970383 | 0.51 |
ENST00000400904.3
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr10_+_60936921 | 0.51 |
ENST00000373878.3
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr18_-_19283649 | 0.51 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr6_-_27114577 | 0.50 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr8_-_67977178 | 0.48 |
ENST00000517736.1
|
COPS5
|
COP9 signalosome subunit 5 |
| chr1_+_207943667 | 0.46 |
ENST00000462968.2
|
CD46
|
CD46 molecule, complement regulatory protein |
| chrX_+_37208521 | 0.45 |
ENST00000378628.4
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr9_+_128510454 | 0.45 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr1_+_219347186 | 0.44 |
ENST00000366928.5
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr6_-_51952418 | 0.43 |
ENST00000371117.3
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr7_+_90338547 | 0.43 |
ENST00000446790.1
|
CDK14
|
cyclin-dependent kinase 14 |
| chr17_-_1395954 | 0.42 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr1_+_64669294 | 0.42 |
ENST00000371077.5
|
UBE2U
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr7_-_121944491 | 0.41 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr1_+_219347203 | 0.41 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr11_-_57194919 | 0.40 |
ENST00000532795.1
|
SLC43A3
|
solute carrier family 43, member 3 |
| chr12_+_4758264 | 0.40 |
ENST00000266544.5
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr3_+_99904531 | 0.39 |
ENST00000429523.2
ENST00000476804.1 |
TMEM30C
|
transmembrane protein 30C |
| chr14_-_21566731 | 0.38 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr22_+_17956618 | 0.38 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr2_+_3800169 | 0.38 |
ENST00000399143.3
|
DCDC2C
|
doublecortin domain containing 2C |
| chr18_-_5197224 | 0.37 |
ENST00000434239.3
|
C18orf42
|
chromosome 18 open reading frame 42 |
| chrX_+_70503037 | 0.37 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr17_+_6544078 | 0.37 |
ENST00000250101.5
|
TXNDC17
|
thioredoxin domain containing 17 |
| chr10_+_24755416 | 0.36 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr22_+_19419425 | 0.36 |
ENST00000333130.3
|
MRPL40
|
mitochondrial ribosomal protein L40 |
| chr8_-_28347737 | 0.36 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr3_-_141747439 | 0.35 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr9_-_73029540 | 0.35 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr5_+_55354822 | 0.35 |
ENST00000511861.1
|
CTD-2227I18.1
|
CTD-2227I18.1 |
| chr7_-_47579188 | 0.34 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr5_+_131705438 | 0.34 |
ENST00000245407.3
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr3_+_147127142 | 0.34 |
ENST00000282928.4
|
ZIC1
|
Zic family member 1 |
| chr1_+_87595433 | 0.34 |
ENST00000469312.2
ENST00000490006.2 |
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chrX_+_70503433 | 0.33 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr6_-_165989936 | 0.33 |
ENST00000354448.4
|
PDE10A
|
phosphodiesterase 10A |
| chr7_+_134576317 | 0.33 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr8_+_94710789 | 0.33 |
ENST00000523475.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr3_-_141747459 | 0.33 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_+_120860695 | 0.33 |
ENST00000505122.2
|
RP11-700N1.1
|
RP11-700N1.1 |
| chr3_-_126327398 | 0.32 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr16_+_29127282 | 0.32 |
ENST00000562902.1
|
RP11-426C22.5
|
RP11-426C22.5 |
| chr4_-_186682716 | 0.31 |
ENST00000445343.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr13_+_24144509 | 0.31 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr11_+_5474638 | 0.31 |
ENST00000341449.2
|
OR51I2
|
olfactory receptor, family 51, subfamily I, member 2 |
| chr1_-_217250231 | 0.30 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr17_+_42925270 | 0.30 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr3_+_159481988 | 0.30 |
ENST00000472451.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr1_-_9953295 | 0.30 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chrX_-_10588595 | 0.29 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr3_+_159481791 | 0.29 |
ENST00000460298.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr17_+_3572087 | 0.29 |
ENST00000248378.5
ENST00000397133.2 |
EMC6
|
ER membrane protein complex subunit 6 |
| chr6_-_8102714 | 0.28 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr11_-_57194817 | 0.28 |
ENST00000529748.1
ENST00000525474.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr2_-_38604398 | 0.28 |
ENST00000443098.1
ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2
|
atlastin GTPase 2 |
| chr17_-_56609302 | 0.28 |
ENST00000581607.1
ENST00000317256.6 ENST00000426861.1 ENST00000580809.1 ENST00000577729.1 ENST00000583291.1 |
SEPT4
|
septin 4 |
| chr4_-_122302163 | 0.28 |
ENST00000394427.2
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr14_+_105331596 | 0.27 |
ENST00000556508.1
ENST00000414716.3 ENST00000453495.1 ENST00000418279.1 |
CEP170B
|
centrosomal protein 170B |
| chr6_+_134758827 | 0.27 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr5_+_125800836 | 0.27 |
ENST00000511134.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr1_-_9970227 | 0.26 |
ENST00000377263.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr17_+_48172639 | 0.26 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr2_+_128848740 | 0.26 |
ENST00000375990.3
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr1_-_93257951 | 0.26 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr12_-_56727676 | 0.26 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr8_-_38326119 | 0.26 |
ENST00000356207.5
ENST00000326324.6 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr2_+_127782787 | 0.26 |
ENST00000564121.1
|
RP11-521O16.2
|
RP11-521O16.2 |
| chrX_-_10588459 | 0.25 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr12_+_49121213 | 0.25 |
ENST00000548380.1
|
LINC00935
|
long intergenic non-protein coding RNA 935 |
| chr6_+_7108210 | 0.25 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr5_-_137667526 | 0.24 |
ENST00000503022.1
|
CDC25C
|
cell division cycle 25C |
| chr7_-_47578840 | 0.24 |
ENST00000450444.1
|
TNS3
|
tensin 3 |
| chr9_-_23779367 | 0.24 |
ENST00000440102.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr10_-_94257512 | 0.23 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr14_+_21458127 | 0.23 |
ENST00000382985.4
ENST00000556670.2 ENST00000553564.1 ENST00000554751.1 ENST00000554283.1 ENST00000555670.1 |
METTL17
|
methyltransferase like 17 |
| chr2_-_99952769 | 0.23 |
ENST00000409434.1
ENST00000434323.1 ENST00000264255.3 |
TXNDC9
|
thioredoxin domain containing 9 |
| chr1_+_198126209 | 0.23 |
ENST00000367383.1
|
NEK7
|
NIMA-related kinase 7 |
| chr7_-_116607526 | 0.23 |
ENST00000420664.1
|
AC106873.4
|
Uncharacterized protein |
| chr7_-_16505440 | 0.23 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr8_-_67874805 | 0.22 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr7_+_107220899 | 0.22 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr2_+_162087577 | 0.22 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr8_-_38326139 | 0.21 |
ENST00000335922.5
ENST00000532791.1 ENST00000397091.5 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr1_-_217311090 | 0.20 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr5_-_137667459 | 0.20 |
ENST00000415130.2
ENST00000356505.3 ENST00000357274.3 ENST00000348983.3 ENST00000323760.6 |
CDC25C
|
cell division cycle 25C |
| chr11_+_66115304 | 0.20 |
ENST00000531602.1
|
RP11-867G23.8
|
Uncharacterized protein |
| chrX_+_37208540 | 0.20 |
ENST00000466533.1
ENST00000542554.1 ENST00000543642.1 ENST00000484460.1 ENST00000449135.2 ENST00000463135.1 ENST00000465127.1 |
PRRG1
TM4SF2
|
proline rich Gla (G-carboxyglutamic acid) 1 Uncharacterized protein; cDNA FLJ59144, highly similar to Tetraspanin-7 |
| chr12_-_53715328 | 0.20 |
ENST00000547757.1
ENST00000394384.3 ENST00000209873.4 |
AAAS
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr1_+_100436065 | 0.19 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chrX_-_69509738 | 0.19 |
ENST00000374454.1
ENST00000239666.4 |
PDZD11
|
PDZ domain containing 11 |
| chr7_+_128379449 | 0.19 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr14_+_45431379 | 0.19 |
ENST00000361577.3
ENST00000361462.2 ENST00000382233.2 |
FAM179B
|
family with sequence similarity 179, member B |
| chr12_-_56727487 | 0.18 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr7_-_22234381 | 0.18 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr7_+_95115210 | 0.18 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr2_+_204193101 | 0.17 |
ENST00000430418.1
ENST00000424558.1 ENST00000261016.6 |
ABI2
|
abl-interactor 2 |
| chr10_-_24911746 | 0.17 |
ENST00000320481.6
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr11_+_57531292 | 0.17 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr4_+_118955635 | 0.17 |
ENST00000433996.2
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr19_-_56826157 | 0.17 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr20_+_59654146 | 0.17 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr11_-_124981475 | 0.17 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr17_-_27621125 | 0.17 |
ENST00000579665.1
ENST00000225388.4 |
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr15_+_75639296 | 0.17 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr14_+_32030582 | 0.17 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMBOX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.6 | 2.4 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.6 | 5.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 1.5 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.4 | 1.8 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.4 | 1.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 1.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.3 | 2.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.3 | 1.3 | GO:0048496 | adherens junction maintenance(GO:0034334) maintenance of organ identity(GO:0048496) |
| 0.3 | 1.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.9 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.2 | 0.7 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.2 | 1.7 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.2 | 1.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 0.9 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.2 | 2.3 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.2 | 1.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 1.6 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 2.1 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.2 | 0.6 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.5 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.2 | 1.4 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.4 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.1 | 0.6 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.7 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 4.8 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.4 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.3 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.1 | 0.8 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 1.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.3 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.1 | 0.6 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 0.8 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.4 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.6 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.1 | 1.8 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 1.9 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 2.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 1.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.6 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.2 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 7.9 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 2.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.2 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 1.1 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.5 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 1.3 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.4 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.6 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 3.7 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 1.1 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 1.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 0.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 1.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.5 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.3 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 1.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.2 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 1.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 1.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 3.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.4 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 4.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.4 | 1.2 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 1.5 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.3 | 1.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 1.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 1.8 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.3 | 9.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.3 | 1.9 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 1.5 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 1.7 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.2 | 0.7 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.2 | 0.7 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.6 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 1.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.6 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 2.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.9 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 2.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 5.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.8 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 1.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 1.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 2.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.8 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 2.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 1.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 3.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.3 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 1.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 3.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 2.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 3.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 5.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 1.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 4.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 5.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |