Project
Illumina Body Map 2
Navigation
Downloads
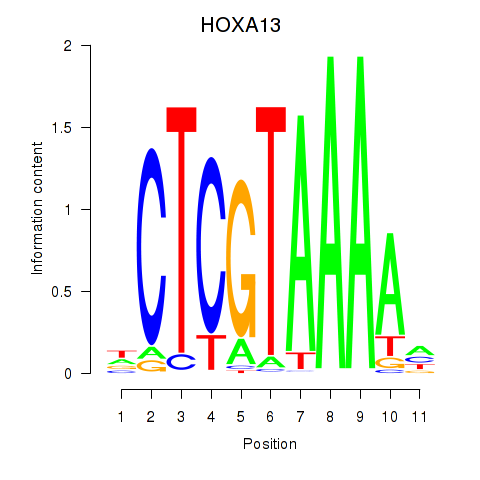
Results for HOXA13
Z-value: 1.31
Transcription factors associated with HOXA13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA13
|
ENSG00000106031.6 | homeobox A13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA13 | hg19_v2_chr7_-_27239703_27239725 | 0.52 | 2.1e-03 | Click! |
Activity profile of HOXA13 motif
Sorted Z-values of HOXA13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_189839046 | 4.70 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr8_-_86290333 | 4.56 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr10_+_24738355 | 4.29 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr1_+_43766642 | 4.06 |
ENST00000372476.3
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr15_-_58571445 | 3.36 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr4_+_70796784 | 3.18 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr1_-_178838404 | 3.12 |
ENST00000444255.1
|
ANGPTL1
|
angiopoietin-like 1 |
| chr14_-_92414294 | 3.11 |
ENST00000554468.1
|
FBLN5
|
fibulin 5 |
| chr4_-_87770416 | 3.02 |
ENST00000273905.6
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
| chr1_+_43766668 | 3.01 |
ENST00000441333.2
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr6_+_19837592 | 2.92 |
ENST00000378700.3
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr10_-_13544945 | 2.90 |
ENST00000378605.3
ENST00000341083.3 |
BEND7
|
BEN domain containing 7 |
| chr11_-_10590238 | 2.89 |
ENST00000256178.3
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr1_+_47533160 | 2.84 |
ENST00000334194.3
|
CYP4Z1
|
cytochrome P450, family 4, subfamily Z, polypeptide 1 |
| chr6_+_88117683 | 2.65 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr7_-_27196267 | 2.60 |
ENST00000242159.3
|
HOXA7
|
homeobox A7 |
| chr19_+_16999654 | 2.59 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr14_-_92414055 | 2.50 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr7_-_16921601 | 2.39 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr7_-_27224795 | 2.39 |
ENST00000006015.3
|
HOXA11
|
homeobox A11 |
| chr16_+_82090028 | 2.30 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr13_-_51101468 | 2.29 |
ENST00000428276.1
|
RP11-175B12.2
|
RP11-175B12.2 |
| chr10_+_118187379 | 2.25 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr20_+_55204351 | 2.25 |
ENST00000201031.2
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr14_-_60952739 | 2.22 |
ENST00000555476.1
ENST00000321731.3 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chr17_-_46688334 | 2.22 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr4_+_128702969 | 2.12 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr6_+_121756809 | 2.01 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr4_-_101111615 | 2.00 |
ENST00000273990.2
|
DDIT4L
|
DNA-damage-inducible transcript 4-like |
| chr3_-_149093499 | 1.99 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr3_-_100712352 | 1.99 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr1_+_13910194 | 1.94 |
ENST00000376057.4
ENST00000510906.1 |
PDPN
|
podoplanin |
| chr2_-_118943930 | 1.94 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr12_-_53074182 | 1.92 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr1_+_166958346 | 1.87 |
ENST00000367872.4
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr15_-_99789736 | 1.84 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr3_+_50211240 | 1.82 |
ENST00000420831.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr11_-_10590118 | 1.77 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr8_-_93029865 | 1.76 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_+_51895621 | 1.75 |
ENST00000333127.3
|
IQCF2
|
IQ motif containing F2 |
| chr19_-_51340469 | 1.73 |
ENST00000326856.4
|
KLK15
|
kallikrein-related peptidase 15 |
| chr12_-_53207842 | 1.72 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr3_+_132316081 | 1.68 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr12_-_91539918 | 1.64 |
ENST00000548218.1
|
DCN
|
decorin |
| chr4_+_15341442 | 1.64 |
ENST00000397700.2
ENST00000295297.4 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr11_+_101983176 | 1.64 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr1_-_147245445 | 1.61 |
ENST00000430508.1
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr8_+_143781513 | 1.60 |
ENST00000292430.6
ENST00000561179.1 ENST00000518841.1 ENST00000519387.1 |
LY6K
|
lymphocyte antigen 6 complex, locus K |
| chr8_-_95274536 | 1.60 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr18_+_31185530 | 1.58 |
ENST00000586327.1
|
ASXL3
|
additional sex combs like 3 (Drosophila) |
| chr7_-_27169801 | 1.57 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr8_+_133879193 | 1.57 |
ENST00000377869.1
ENST00000220616.4 |
TG
|
thyroglobulin |
| chr1_-_46642154 | 1.54 |
ENST00000540385.1
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr14_-_80678512 | 1.51 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr4_+_54243917 | 1.51 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr4_+_42399856 | 1.51 |
ENST00000319234.4
|
SHISA3
|
shisa family member 3 |
| chr12_+_12878829 | 1.49 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr12_-_102874102 | 1.45 |
ENST00000392905.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr17_+_77030267 | 1.42 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr2_+_192109911 | 1.41 |
ENST00000418908.1
ENST00000339514.4 ENST00000392318.3 |
MYO1B
|
myosin IB |
| chr3_-_99569821 | 1.36 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr11_+_113185778 | 1.34 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr17_-_17480779 | 1.34 |
ENST00000395782.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr8_+_35649365 | 1.31 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr3_-_126373929 | 1.30 |
ENST00000523403.1
ENST00000524230.2 |
TXNRD3
|
thioredoxin reductase 3 |
| chr7_-_73256838 | 1.28 |
ENST00000297873.4
|
WBSCR27
|
Williams Beuren syndrome chromosome region 27 |
| chr3_-_51909600 | 1.28 |
ENST00000446461.1
|
IQCF5
|
IQ motif containing F5 |
| chr2_-_23747214 | 1.25 |
ENST00000430988.1
|
AC011239.1
|
Uncharacterized protein |
| chr1_+_78383813 | 1.24 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr2_+_207804278 | 1.24 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr7_+_23749945 | 1.23 |
ENST00000354639.3
ENST00000531170.1 ENST00000444333.2 ENST00000428484.1 |
STK31
|
serine/threonine kinase 31 |
| chr12_+_120105558 | 1.20 |
ENST00000229328.5
ENST00000541640.1 |
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr15_+_59908633 | 1.19 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chrX_+_35816459 | 1.18 |
ENST00000399988.1
ENST00000399992.1 ENST00000399987.1 ENST00000399989.1 |
MAGEB16
|
melanoma antigen family B, 16 |
| chr15_-_55881135 | 1.16 |
ENST00000302000.6
|
PYGO1
|
pygopus family PHD finger 1 |
| chr1_+_78511586 | 1.15 |
ENST00000370759.3
|
GIPC2
|
GIPC PDZ domain containing family, member 2 |
| chr4_-_110723134 | 1.15 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr3_-_123339418 | 1.13 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr19_+_852291 | 1.13 |
ENST00000263621.1
|
ELANE
|
elastase, neutrophil expressed |
| chr19_+_859654 | 1.12 |
ENST00000592860.1
|
CFD
|
complement factor D (adipsin) |
| chr16_+_82068585 | 1.12 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr17_-_15496722 | 1.11 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chrX_+_37850026 | 1.11 |
ENST00000341016.3
|
CXorf27
|
chromosome X open reading frame 27 |
| chr5_-_146781153 | 1.11 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr1_+_78769549 | 1.10 |
ENST00000370758.1
|
PTGFR
|
prostaglandin F receptor (FP) |
| chr7_+_23749767 | 1.05 |
ENST00000355870.3
|
STK31
|
serine/threonine kinase 31 |
| chr11_+_7110165 | 1.04 |
ENST00000306904.5
|
RBMXL2
|
RNA binding motif protein, X-linked-like 2 |
| chr3_-_123339343 | 1.04 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr11_+_62037622 | 1.04 |
ENST00000227918.2
ENST00000525380.1 |
SCGB2A2
|
secretoglobin, family 2A, member 2 |
| chr11_+_26495447 | 1.03 |
ENST00000531568.1
|
ANO3
|
anoctamin 3 |
| chr3_+_108855558 | 1.03 |
ENST00000467240.1
ENST00000477643.1 ENST00000479039.1 ENST00000593799.1 |
RP11-59E19.1
|
RP11-59E19.1 |
| chr17_+_55183261 | 1.02 |
ENST00000576295.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr7_+_23749894 | 1.01 |
ENST00000433467.2
|
STK31
|
serine/threonine kinase 31 |
| chr1_+_199996733 | 1.01 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr3_-_100712292 | 1.01 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr7_+_80275953 | 1.00 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr19_+_50084561 | 0.99 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr7_+_80275621 | 0.98 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr3_+_154801678 | 0.97 |
ENST00000462837.1
|
MME
|
membrane metallo-endopeptidase |
| chr10_-_113943447 | 0.97 |
ENST00000369425.1
ENST00000348367.4 ENST00000423155.1 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr16_-_54304608 | 0.95 |
ENST00000561336.1
|
RP11-324D17.1
|
HCG2045435; Uncharacterized protein |
| chr7_-_69062391 | 0.93 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr9_+_67977438 | 0.92 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr1_+_152730499 | 0.92 |
ENST00000368773.1
|
KPRP
|
keratinocyte proline-rich protein |
| chr13_-_24895566 | 0.92 |
ENST00000422229.2
|
AL359736.1
|
protein PCOTH isoform 1 |
| chr1_-_153085984 | 0.92 |
ENST00000468739.1
|
SPRR2F
|
small proline-rich protein 2F |
| chr9_+_706842 | 0.91 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr7_+_23749854 | 0.90 |
ENST00000456014.2
|
STK31
|
serine/threonine kinase 31 |
| chr1_-_204183071 | 0.89 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr7_+_80275663 | 0.89 |
ENST00000413265.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr6_+_148663729 | 0.89 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr11_+_60995849 | 0.89 |
ENST00000537932.1
|
PGA4
|
pepsinogen 4, group I (pepsinogen A) |
| chr7_+_80275752 | 0.89 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr3_+_111578027 | 0.88 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr2_+_33359687 | 0.88 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr20_-_44420507 | 0.84 |
ENST00000243938.4
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr2_+_170366203 | 0.84 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr19_-_7167989 | 0.84 |
ENST00000600492.1
|
INSR
|
insulin receptor |
| chr7_+_23749822 | 0.83 |
ENST00000422637.1
|
STK31
|
serine/threonine kinase 31 |
| chr9_+_6215799 | 0.82 |
ENST00000417746.2
ENST00000456383.2 |
IL33
|
interleukin 33 |
| chr2_+_33359646 | 0.82 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_+_199996702 | 0.82 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr19_+_48497901 | 0.82 |
ENST00000339841.2
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr1_+_145525015 | 0.81 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr1_+_19967014 | 0.81 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr3_-_112013071 | 0.79 |
ENST00000487372.1
ENST00000486574.1 ENST00000305815.5 |
SLC9C1
|
solute carrier family 9, subfamily C (Na+-transporting carboxylic acid decarboxylase), member 1 |
| chr10_-_50970322 | 0.77 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr1_-_209975494 | 0.77 |
ENST00000456314.1
|
IRF6
|
interferon regulatory factor 6 |
| chr12_+_26274917 | 0.76 |
ENST00000538142.1
|
SSPN
|
sarcospan |
| chr8_-_16050214 | 0.76 |
ENST00000262101.5
|
MSR1
|
macrophage scavenger receptor 1 |
| chr15_+_74165945 | 0.75 |
ENST00000535547.2
ENST00000300504.2 ENST00000562056.1 |
TBC1D21
|
TBC1 domain family, member 21 |
| chr19_-_51893827 | 0.75 |
ENST00000574814.1
|
CTD-2616J11.4
|
chromosome 19 open reading frame 84 |
| chr14_+_24540046 | 0.75 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr8_-_16050288 | 0.75 |
ENST00000350896.3
|
MSR1
|
macrophage scavenger receptor 1 |
| chr10_+_102756800 | 0.73 |
ENST00000370223.3
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr1_+_202431859 | 0.70 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr15_+_69373184 | 0.70 |
ENST00000558147.1
ENST00000440444.1 |
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr8_+_118147498 | 0.70 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr3_-_160823158 | 0.69 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr12_-_56367101 | 0.69 |
ENST00000549233.2
|
PMEL
|
premelanosome protein |
| chr12_-_91546926 | 0.68 |
ENST00000550758.1
|
DCN
|
decorin |
| chr10_-_50143242 | 0.68 |
ENST00000298124.3
|
LRRC18
|
leucine rich repeat containing 18 |
| chrX_-_33357558 | 0.68 |
ENST00000288447.4
|
DMD
|
dystrophin |
| chr21_-_26797019 | 0.67 |
ENST00000440205.1
|
LINC00158
|
long intergenic non-protein coding RNA 158 |
| chr4_-_82136114 | 0.67 |
ENST00000395578.1
ENST00000418486.2 |
PRKG2
|
protein kinase, cGMP-dependent, type II |
| chr12_-_8693469 | 0.67 |
ENST00000545274.1
ENST00000446457.2 |
CLEC4E
|
C-type lectin domain family 4, member E |
| chr4_+_94125100 | 0.67 |
ENST00000512631.1
|
GRID2
|
glutamate receptor, ionotropic, delta 2 |
| chr6_+_155334780 | 0.67 |
ENST00000538270.1
ENST00000535231.1 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr15_+_69373210 | 0.67 |
ENST00000435479.1
ENST00000559870.1 |
LINC00277
RP11-809H16.5
|
long intergenic non-protein coding RNA 277 RP11-809H16.5 |
| chr3_-_160823040 | 0.66 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr4_+_69313145 | 0.66 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr11_+_94706804 | 0.66 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr1_-_91182794 | 0.66 |
ENST00000370445.4
|
BARHL2
|
BarH-like homeobox 2 |
| chr9_+_112887772 | 0.65 |
ENST00000259318.7
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr12_-_50677255 | 0.65 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr5_-_124081008 | 0.65 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr19_-_39694894 | 0.64 |
ENST00000318438.6
|
SYCN
|
syncollin |
| chrX_-_11284095 | 0.64 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr10_+_106028923 | 0.63 |
ENST00000338595.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr12_-_71182695 | 0.62 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr6_-_51952418 | 0.62 |
ENST00000371117.3
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr11_-_4719072 | 0.61 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr7_-_50633078 | 0.61 |
ENST00000444124.2
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr19_+_36545781 | 0.60 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chr3_-_150996146 | 0.60 |
ENST00000494668.1
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr3_+_113667354 | 0.59 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr11_+_101785727 | 0.58 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr15_+_72690614 | 0.58 |
ENST00000341689.3
ENST00000567679.1 |
TMEM202
|
transmembrane protein 202 |
| chr4_-_147866960 | 0.57 |
ENST00000513335.1
|
TTC29
|
tetratricopeptide repeat domain 29 |
| chr14_+_23654525 | 0.57 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr8_-_30706608 | 0.57 |
ENST00000256246.2
|
TEX15
|
testis expressed 15 |
| chr1_-_112106578 | 0.57 |
ENST00000369717.4
|
ADORA3
|
adenosine A3 receptor |
| chr16_-_21289627 | 0.57 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr12_+_121163602 | 0.56 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr12_+_51317788 | 0.56 |
ENST00000550502.1
|
METTL7A
|
methyltransferase like 7A |
| chr12_+_45609893 | 0.55 |
ENST00000320560.8
|
ANO6
|
anoctamin 6 |
| chr1_+_222886694 | 0.54 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr9_-_21305312 | 0.53 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr4_+_190802670 | 0.53 |
ENST00000503609.1
|
RP11-463J17.1
|
RP11-463J17.1 |
| chr4_-_89978299 | 0.53 |
ENST00000511976.1
ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr19_+_49496782 | 0.53 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr11_+_61015594 | 0.52 |
ENST00000451616.2
|
PGA5
|
pepsinogen 5, group I (pepsinogen A) |
| chr4_-_159956333 | 0.52 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr3_+_160117062 | 0.52 |
ENST00000497311.1
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr3_+_154801312 | 0.51 |
ENST00000497890.1
|
MME
|
membrane metallo-endopeptidase |
| chr8_+_94710789 | 0.51 |
ENST00000523475.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr7_-_75401513 | 0.51 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr3_-_160822858 | 0.51 |
ENST00000488170.1
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr13_-_36429763 | 0.51 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr17_+_11501748 | 0.50 |
ENST00000262442.4
ENST00000579828.1 |
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr2_+_33359473 | 0.50 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr18_+_61223393 | 0.50 |
ENST00000269491.1
ENST00000382768.1 |
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr1_-_31845914 | 0.49 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr20_+_59654146 | 0.47 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr12_-_12491608 | 0.47 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr7_-_91808441 | 0.47 |
ENST00000437357.1
ENST00000458448.1 |
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chr6_+_151042224 | 0.47 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr2_+_7073174 | 0.46 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr3_+_129159039 | 0.46 |
ENST00000507564.1
ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr4_+_106631966 | 0.46 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.8 | 6.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.6 | 1.9 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.6 | 2.9 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.4 | 5.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.4 | 1.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 2.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.3 | 3.4 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.3 | 1.3 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.3 | 3.8 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.3 | 0.9 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 1.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.3 | 1.4 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.2 | 0.7 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.2 | 1.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 1.0 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.2 | 0.6 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.2 | 0.6 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.2 | 2.6 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 2.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 1.1 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.2 | 1.4 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 2.6 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.2 | 0.8 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.2 | 0.8 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.2 | 0.8 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.2 | 2.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 3.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 4.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.4 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 1.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 1.8 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 1.6 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 1.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 2.5 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.1 | 7.2 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 1.5 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 1.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 1.8 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.4 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 | 0.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.7 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 1.5 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 0.5 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.6 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.6 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.4 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.7 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 2.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.3 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.1 | 1.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 1.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.2 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 1.9 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 3.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.9 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.6 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.8 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.6 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 1.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.8 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.7 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.6 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.7 | GO:0035588 | G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 1.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.3 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 1.0 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.2 | GO:0010193 | response to ozone(GO:0010193) response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.3 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.0 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.0 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 1.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.4 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 1.3 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 1.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.7 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.5 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 1.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.2 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.4 | GO:0007350 | blastoderm segmentation(GO:0007350) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.5 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.9 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.6 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 1.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.6 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.7 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.6 | GO:0040008 | regulation of growth(GO:0040008) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.8 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.5 | GO:0032259 | methylation(GO:0032259) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.5 | 1.9 | GO:0030849 | autosome(GO:0030849) |
| 0.3 | 1.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 4.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.2 | 0.8 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.2 | 2.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 3.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.4 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 1.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 4.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 3.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 2.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 5.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 1.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.8 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 2.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 3.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.4 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.6 | 4.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 1.6 | GO:0086079 | gap junction channel activity involved in atrial cardiac muscle cell-AV node cell electrical coupling(GO:0086076) gap junction channel activity involved in bundle of His cell-Purkinje myocyte electrical coupling(GO:0086078) gap junction channel activity involved in Purkinje myocyte-ventricular cardiac muscle cell electrical coupling(GO:0086079) |
| 0.5 | 1.9 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.4 | 1.3 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.4 | 1.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.4 | 2.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.4 | 1.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.4 | 2.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 3.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.3 | 2.0 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 2.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 4.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 2.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 3.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 1.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.2 | 0.5 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.2 | 0.3 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.6 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 1.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.9 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 4.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.8 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 2.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 3.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.8 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.9 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 1.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 1.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 2.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 1.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 7.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 6.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 1.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.7 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 7.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 2.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 4.8 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 4.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 4.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 4.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 3.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 10.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 3.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 2.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 4.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 2.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 4.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 1.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 2.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 2.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 4.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |