Project
Illumina Body Map 2
Navigation
Downloads
Results for HOXB7
Z-value: 1.42
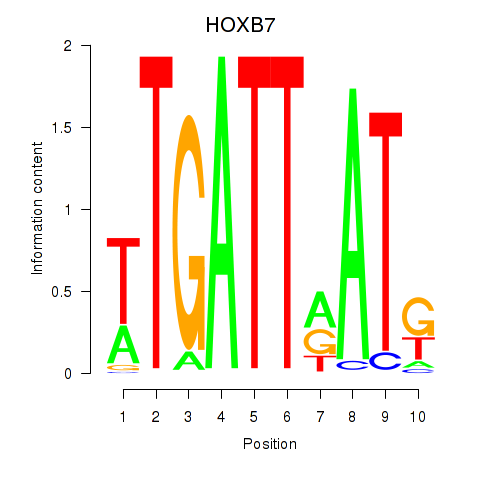
Transcription factors associated with HOXB7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB7
|
ENSG00000260027.3 | homeobox B7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB7 | hg19_v2_chr17_-_46688334_46688385 | -0.01 | 9.7e-01 | Click! |
Activity profile of HOXB7 motif
Sorted Z-values of HOXB7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_101988627 | 8.37 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr12_+_101988774 | 8.17 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr10_+_69865866 | 6.22 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr12_-_22063787 | 4.31 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr2_+_234580525 | 4.14 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr3_-_46608010 | 3.94 |
ENST00000395905.3
|
LRRC2
|
leucine rich repeat containing 2 |
| chr2_+_234580499 | 3.61 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr15_-_93616340 | 3.24 |
ENST00000557420.1
ENST00000542321.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr10_+_69869237 | 3.17 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr10_+_24755416 | 3.16 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr2_+_168043793 | 3.15 |
ENST00000409273.1
ENST00000409605.1 |
XIRP2
|
xin actin-binding repeat containing 2 |
| chr3_+_35682913 | 3.00 |
ENST00000449196.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr4_-_186570679 | 2.99 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_-_85882145 | 2.93 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr12_-_47226152 | 2.91 |
ENST00000546940.1
|
SLC38A4
|
solute carrier family 38, member 4 |
| chr8_-_91095099 | 2.90 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr10_+_13142225 | 2.69 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr4_-_72649763 | 2.65 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr10_+_13141585 | 2.59 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr6_+_54173227 | 2.56 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr13_-_48612067 | 2.56 |
ENST00000543413.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr3_+_158787041 | 2.52 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr10_+_13142075 | 2.49 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr2_+_234621551 | 2.49 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr20_+_238357 | 2.47 |
ENST00000382376.3
|
DEFB132
|
defensin, beta 132 |
| chrX_+_130192318 | 2.37 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr4_-_44653636 | 2.33 |
ENST00000415895.4
ENST00000332990.5 |
YIPF7
|
Yip1 domain family, member 7 |
| chr8_-_109799793 | 2.33 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr12_+_21679220 | 2.31 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr16_+_7560114 | 2.29 |
ENST00000570626.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_+_234637754 | 2.25 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr18_+_29171689 | 2.25 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr11_-_2162162 | 2.21 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr4_+_69962185 | 2.14 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr15_-_93616892 | 2.08 |
ENST00000556658.1
ENST00000538818.1 ENST00000425933.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr3_+_189349162 | 2.06 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr14_-_23624511 | 2.03 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr3_+_148415571 | 2.03 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr4_+_110834033 | 2.03 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr8_-_67874805 | 2.02 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr3_-_196242233 | 1.99 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr4_+_69962212 | 1.97 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr11_-_89223883 | 1.97 |
ENST00000528341.1
|
NOX4
|
NADPH oxidase 4 |
| chr9_+_74729511 | 1.96 |
ENST00000545168.1
|
GDA
|
guanine deaminase |
| chr3_-_58613323 | 1.94 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr4_-_100212132 | 1.93 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr3_+_159557637 | 1.91 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr15_+_48483736 | 1.89 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr1_-_216896674 | 1.88 |
ENST00000475275.1
ENST00000469486.1 ENST00000481543.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr12_-_103344615 | 1.85 |
ENST00000546844.1
|
PAH
|
phenylalanine hydroxylase |
| chr8_-_42360015 | 1.85 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr4_-_111558135 | 1.84 |
ENST00000394598.2
ENST00000394595.3 |
PITX2
|
paired-like homeodomain 2 |
| chr4_-_140005341 | 1.80 |
ENST00000379549.2
ENST00000512627.1 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr1_-_216896780 | 1.80 |
ENST00000459955.1
ENST00000366937.1 ENST00000408911.3 ENST00000391890.3 |
ESRRG
|
estrogen-related receptor gamma |
| chrX_+_130192216 | 1.79 |
ENST00000276211.5
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr3_+_173116225 | 1.78 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr4_-_69817481 | 1.76 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr12_+_81471816 | 1.73 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr3_+_189507523 | 1.73 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr12_+_54402790 | 1.73 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr16_+_15596123 | 1.72 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr4_+_113739244 | 1.67 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr16_-_20364030 | 1.67 |
ENST00000396134.2
ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD
|
uromodulin |
| chr4_+_71600063 | 1.67 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr11_+_64358686 | 1.67 |
ENST00000473690.1
|
SLC22A12
|
solute carrier family 22 (organic anion/urate transporter), member 12 |
| chr3_+_148545586 | 1.66 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr2_-_142888573 | 1.65 |
ENST00000434794.1
|
LRP1B
|
low density lipoprotein receptor-related protein 1B |
| chr3_-_47932759 | 1.60 |
ENST00000441748.2
ENST00000335271.5 |
MAP4
|
microtubule-associated protein 4 |
| chr7_-_120498357 | 1.60 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr4_-_118006697 | 1.59 |
ENST00000310754.4
|
TRAM1L1
|
translocation associated membrane protein 1-like 1 |
| chr17_+_67498295 | 1.59 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr8_-_70747205 | 1.58 |
ENST00000260126.4
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr4_+_69681710 | 1.57 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr4_-_70080449 | 1.57 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr12_-_91573249 | 1.56 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr2_+_169659121 | 1.55 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr6_-_51952418 | 1.55 |
ENST00000371117.3
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr19_-_30199516 | 1.53 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr10_+_13141441 | 1.52 |
ENST00000263036.5
|
OPTN
|
optineurin |
| chr9_-_73736511 | 1.51 |
ENST00000377110.3
ENST00000377111.2 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr7_+_15728003 | 1.49 |
ENST00000442176.1
|
AC005550.4
|
AC005550.4 |
| chr9_+_140445651 | 1.49 |
ENST00000371443.5
|
MRPL41
|
mitochondrial ribosomal protein L41 |
| chr3_-_155011483 | 1.49 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr3_-_33686925 | 1.48 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr16_-_51185172 | 1.47 |
ENST00000251020.4
|
SALL1
|
spalt-like transcription factor 1 |
| chr3_+_35683651 | 1.46 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr4_+_108925787 | 1.45 |
ENST00000454409.2
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr4_-_140005443 | 1.45 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr4_+_144354644 | 1.44 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr17_-_37353950 | 1.44 |
ENST00000394310.3
ENST00000394303.3 ENST00000344140.5 |
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr12_-_91573132 | 1.43 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr11_+_64358722 | 1.43 |
ENST00000336464.7
|
SLC22A12
|
solute carrier family 22 (organic anion/urate transporter), member 12 |
| chr2_+_234590556 | 1.42 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr2_+_169658928 | 1.41 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr2_+_27886330 | 1.40 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr13_-_33760216 | 1.40 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr16_-_20364122 | 1.39 |
ENST00000396138.4
ENST00000577168.1 |
UMOD
|
uromodulin |
| chr6_-_110736742 | 1.35 |
ENST00000368924.3
ENST00000368923.3 |
DDO
|
D-aspartate oxidase |
| chr1_-_165414414 | 1.33 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr3_+_148508845 | 1.31 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr2_+_234627424 | 1.30 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr7_+_30185406 | 1.29 |
ENST00000324489.5
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr3_+_2933893 | 1.28 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr9_-_129885010 | 1.28 |
ENST00000373425.3
|
ANGPTL2
|
angiopoietin-like 2 |
| chr2_+_234526272 | 1.27 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr4_-_48014931 | 1.26 |
ENST00000420489.2
ENST00000504722.1 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr2_+_27237615 | 1.26 |
ENST00000458529.1
ENST00000402218.1 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr19_+_13135386 | 1.25 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr9_+_12693336 | 1.24 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr14_+_55034599 | 1.24 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr16_-_21289627 | 1.24 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr3_+_156009623 | 1.24 |
ENST00000389634.5
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr8_+_12803146 | 1.23 |
ENST00000447063.2
|
KIAA1456
|
KIAA1456 |
| chr5_-_78809950 | 1.23 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chrM_+_9207 | 1.23 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr3_+_189507432 | 1.23 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr7_+_30185496 | 1.22 |
ENST00000455738.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr10_-_62332357 | 1.22 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr15_-_54025300 | 1.21 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr3_-_169587621 | 1.20 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr6_+_50786414 | 1.20 |
ENST00000344788.3
ENST00000393655.3 ENST00000263046.4 |
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr17_+_58018269 | 1.20 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr9_-_93405352 | 1.20 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr19_+_12203100 | 1.18 |
ENST00000596883.1
|
ZNF788
|
zinc finger family member 788 |
| chr21_+_44313375 | 1.17 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr2_+_234600253 | 1.14 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr5_-_125930877 | 1.14 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr16_-_51185149 | 1.14 |
ENST00000566102.1
ENST00000541611.1 |
SALL1
|
spalt-like transcription factor 1 |
| chr8_-_66750978 | 1.13 |
ENST00000523253.1
|
PDE7A
|
phosphodiesterase 7A |
| chr16_-_63651939 | 1.13 |
ENST00000563855.1
ENST00000568932.1 ENST00000564290.1 |
RP11-368L12.1
|
RP11-368L12.1 |
| chr9_-_107361788 | 1.13 |
ENST00000374779.2
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5 |
| chr11_-_123525648 | 1.13 |
ENST00000527836.1
|
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr17_+_67498538 | 1.13 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr5_-_59481406 | 1.12 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr15_+_54901540 | 1.12 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr3_+_35722487 | 1.11 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr11_-_63376013 | 1.11 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr6_+_25754927 | 1.10 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr17_+_21030260 | 1.09 |
ENST00000579303.1
|
DHRS7B
|
dehydrogenase/reductase (SDR family) member 7B |
| chr4_-_152329987 | 1.08 |
ENST00000508847.1
|
RP11-610P16.1
|
RP11-610P16.1 |
| chr17_+_22022437 | 1.08 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr5_+_125695805 | 1.07 |
ENST00000513040.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr11_-_4389616 | 1.07 |
ENST00000408920.2
|
OR52B4
|
olfactory receptor, family 52, subfamily B, member 4 |
| chr11_-_123525289 | 1.06 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr17_-_10701241 | 1.05 |
ENST00000578763.1
|
LINC00675
|
long intergenic non-protein coding RNA 675 |
| chr16_+_82660560 | 1.05 |
ENST00000268613.10
ENST00000565636.1 ENST00000431540.3 ENST00000428848.3 |
CDH13
|
cadherin 13 |
| chr8_+_68864330 | 1.05 |
ENST00000288368.4
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr11_-_5345582 | 1.04 |
ENST00000328813.2
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2 |
| chr7_-_96132835 | 1.04 |
ENST00000356686.1
|
C7orf76
|
chromosome 7 open reading frame 76 |
| chr12_+_80750652 | 1.04 |
ENST00000550182.1
|
OTOGL
|
otogelin-like |
| chr21_-_31588365 | 1.03 |
ENST00000399899.1
|
CLDN8
|
claudin 8 |
| chr1_-_209957882 | 1.03 |
ENST00000294811.1
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr4_-_186734275 | 1.00 |
ENST00000456060.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_178103144 | 1.00 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr2_-_200322723 | 0.97 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr3_+_35722424 | 0.97 |
ENST00000396481.2
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_40141502 | 0.97 |
ENST00000539167.1
|
MYRIP
|
myosin VIIA and Rab interacting protein |
| chrX_-_19504642 | 0.96 |
ENST00000469203.2
|
MAP3K15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr18_+_44526744 | 0.95 |
ENST00000585469.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr11_-_47664072 | 0.94 |
ENST00000542981.1
ENST00000530428.1 ENST00000302503.3 |
MTCH2
|
mitochondrial carrier 2 |
| chr12_-_71031185 | 0.94 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr8_+_12803176 | 0.93 |
ENST00000524591.2
|
KIAA1456
|
KIAA1456 |
| chr6_-_51952367 | 0.92 |
ENST00000340994.4
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr4_+_144106063 | 0.92 |
ENST00000510377.1
|
USP38
|
ubiquitin specific peptidase 38 |
| chr16_+_15489603 | 0.91 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr13_-_110438914 | 0.91 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr17_-_42992856 | 0.91 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr18_+_28923589 | 0.90 |
ENST00000462981.2
|
DSG1
|
desmoglein 1 |
| chr8_-_110986918 | 0.89 |
ENST00000297404.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr8_+_98788057 | 0.89 |
ENST00000517924.1
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chrM_+_10053 | 0.89 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr1_-_112106578 | 0.88 |
ENST00000369717.4
|
ADORA3
|
adenosine A3 receptor |
| chr2_+_234545148 | 0.88 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chrX_+_114827818 | 0.88 |
ENST00000420625.2
|
PLS3
|
plastin 3 |
| chr12_+_123944070 | 0.88 |
ENST00000412157.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr12_+_8309630 | 0.88 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr3_-_122712079 | 0.88 |
ENST00000393583.2
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr5_-_38557561 | 0.88 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr7_+_20370746 | 0.87 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr16_-_70835034 | 0.87 |
ENST00000261776.5
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr14_-_94789663 | 0.87 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr10_-_18948208 | 0.85 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr9_-_73029540 | 0.85 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr3_+_63428752 | 0.84 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr14_-_74417096 | 0.84 |
ENST00000286544.3
|
FAM161B
|
family with sequence similarity 161, member B |
| chr12_-_11339543 | 0.84 |
ENST00000334266.1
|
TAS2R42
|
taste receptor, type 2, member 42 |
| chr4_+_70146217 | 0.84 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr5_-_88120151 | 0.84 |
ENST00000506716.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr4_+_144106080 | 0.83 |
ENST00000307017.4
|
USP38
|
ubiquitin specific peptidase 38 |
| chr2_+_120303717 | 0.83 |
ENST00000594371.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr5_-_127418573 | 0.82 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr1_-_3713042 | 0.82 |
ENST00000378251.1
|
LRRC47
|
leucine rich repeat containing 47 |
| chr12_-_91573316 | 0.82 |
ENST00000393155.1
|
DCN
|
decorin |
| chr7_+_116654958 | 0.82 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr5_-_125930929 | 0.82 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr3_-_187388173 | 0.81 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr1_-_178840157 | 0.80 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr12_+_20968608 | 0.80 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr6_+_29068386 | 0.79 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor, family 2, subfamily J, member 1 (gene/pseudogene) |
| chr2_+_44001172 | 0.79 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 10.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 1.4 | 5.6 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 1.3 | 1.3 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 1.0 | 9.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.0 | 7.1 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.8 | 16.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.7 | 2.0 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.7 | 3.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.6 | 2.6 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.6 | 3.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.5 | 0.5 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.5 | 9.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.5 | 2.0 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.5 | 2.0 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.5 | 2.9 | GO:0072025 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.5 | 5.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.5 | 2.3 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.4 | 1.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.4 | 1.8 | GO:0097115 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.4 | 2.6 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.4 | 1.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 | 1.2 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.4 | 1.8 | GO:0035993 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.4 | 1.8 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 1.7 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.3 | 3.2 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.3 | 1.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.3 | 0.6 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.3 | 5.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.2 | 3.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 3.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 1.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 1.6 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 1.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 1.4 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 2.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 1.7 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.2 | 1.2 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 2.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 0.8 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.2 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.2 | 0.9 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.2 | 3.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 0.9 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 | 0.7 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.2 | 0.5 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.2 | 0.5 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.2 | 1.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 0.7 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 0.8 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 0.8 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 1.0 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 2.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.6 | GO:2001248 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.6 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 2.0 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.4 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 1.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.4 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.7 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.8 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.8 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 1.2 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.1 | 0.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.5 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.1 | 0.5 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 1.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.7 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 1.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.9 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 1.7 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.4 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 1.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.5 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 0.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 1.8 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.3 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.7 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.8 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.8 | GO:0009410 | xenobiotic metabolic process(GO:0006805) response to xenobiotic stimulus(GO:0009410) cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 0.9 | GO:0015911 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 4.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 0.7 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.8 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 2.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.3 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.3 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 1.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.3 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) swimming behavior(GO:0036269) |
| 0.1 | 0.2 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 4.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.7 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 4.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.3 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 1.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 3.3 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 0.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.5 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 2.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.4 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 0.6 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.3 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.2 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.1 | 2.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 2.0 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 1.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 1.8 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.8 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 2.5 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 1.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.6 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.6 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.9 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 3.0 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 1.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 2.5 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.8 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.7 | GO:1902083 | cellular response to hydroperoxide(GO:0071447) negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 1.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 2.7 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 0.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.5 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 1.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.6 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 6.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.4 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.7 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.3 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.9 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 2.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.9 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.9 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.9 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.3 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.5 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 2.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.7 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 2.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 4.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.5 | GO:0048869 | cellular developmental process(GO:0048869) |
| 0.0 | 0.6 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.4 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 0.1 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 2.8 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 1.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.5 | 15.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.4 | 10.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 0.9 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.2 | 2.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 3.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.8 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.2 | 0.8 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 1.6 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 0.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.6 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 3.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.9 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.4 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 9.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.5 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.5 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 2.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.8 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 1.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 2.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 2.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.5 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 19.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 2.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.6 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 6.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 3.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 2.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 3.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 4.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 2.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 1.2 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 6.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 2.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 2.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 10.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 2.3 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 1.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.7 | GO:0034703 | cation channel complex(GO:0034703) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.9 | 2.6 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.7 | 25.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.7 | 2.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.7 | 2.0 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.6 | 2.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.6 | 1.9 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.5 | 2.6 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.5 | 27.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.5 | 3.7 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.4 | 2.3 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.4 | 1.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.4 | 5.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.3 | 1.7 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.3 | 1.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 5.6 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.3 | 1.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.3 | 1.8 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.3 | 3.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.3 | 0.8 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 2.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 2.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 9.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 1.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.2 | 0.9 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 1.7 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 1.8 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 2.6 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.2 | 0.8 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.2 | 3.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 1.7 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 3.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.0 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.6 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 2.0 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.8 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 0.6 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 3.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.5 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 1.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.4 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 1.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.3 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 3.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.3 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.1 | 0.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.7 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 3.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.3 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 3.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 3.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.0 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 1.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.3 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 1.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 2.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 1.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 2.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 2.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 2.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 3.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 1.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 1.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 3.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 3.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.7 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 2.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.8 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 2.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 2.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.9 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 5.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.4 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 4.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.9 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 6.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 6.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 4.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 3.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 25.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 16.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 3.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 4.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 4.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 2.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 5.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 3.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 0.9 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 3.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 2.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.7 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 5.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 2.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 2.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 3.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 4.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 4.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.0 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.7 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.5 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |