Project
Illumina Body Map 2
Navigation
Downloads
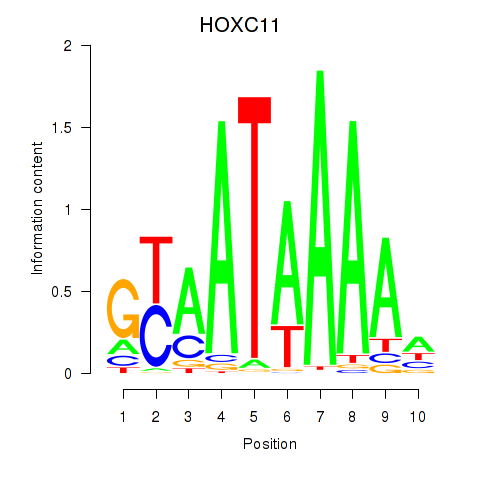
Results for HOXC11
Z-value: 1.50
Transcription factors associated with HOXC11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC11
|
ENSG00000123388.4 | homeobox C11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC11 | hg19_v2_chr12_+_54366894_54366922 | 0.11 | 5.4e-01 | Click! |
Activity profile of HOXC11 motif
Sorted Z-values of HOXC11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_104831472 | 5.60 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr12_+_54384370 | 4.30 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr2_-_190044480 | 4.24 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr5_-_111091948 | 4.21 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr17_-_38821373 | 4.07 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr1_-_204135450 | 3.93 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr8_+_76452097 | 3.90 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr12_+_52203789 | 3.66 |
ENST00000599343.1
|
AC068987.1
|
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
| chrX_+_105192423 | 3.45 |
ENST00000540278.1
|
NRK
|
Nik related kinase |
| chr10_-_96829246 | 3.38 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr1_+_110993795 | 3.32 |
ENST00000271331.3
|
PROK1
|
prokineticin 1 |
| chr8_+_134203273 | 3.02 |
ENST00000250160.6
|
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr2_+_166428839 | 2.84 |
ENST00000342316.4
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr18_+_68002675 | 2.83 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr4_+_95972822 | 2.68 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr8_+_134203303 | 2.63 |
ENST00000519433.1
ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr22_+_41956767 | 2.60 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr3_+_164924716 | 2.50 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr1_+_209878182 | 2.37 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr6_+_88054530 | 2.34 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr4_-_87028478 | 2.32 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr8_+_104831440 | 2.27 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr19_-_10420459 | 2.26 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr20_+_58179582 | 2.23 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr2_+_228678550 | 2.21 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr12_-_96184533 | 2.14 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr7_+_134464414 | 2.07 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr15_-_83952071 | 2.06 |
ENST00000569704.1
|
BNC1
|
basonuclin 1 |
| chr8_-_71157595 | 2.05 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr4_-_174451370 | 2.05 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr8_+_104831554 | 2.04 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr19_+_46367518 | 2.03 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr7_+_134464376 | 1.99 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr20_+_52105495 | 1.98 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr12_-_94673956 | 1.95 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr7_+_35756400 | 1.94 |
ENST00000585952.1
|
AC018647.3
|
AC018647.3 |
| chr5_-_115872124 | 1.92 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chrX_-_10851762 | 1.90 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr6_+_108487245 | 1.83 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr5_-_34043310 | 1.82 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr7_+_35756092 | 1.81 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr4_+_169013666 | 1.76 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr1_+_199996702 | 1.75 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr14_+_101295638 | 1.73 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr19_-_19626838 | 1.66 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr1_+_209859510 | 1.64 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr12_-_96184913 | 1.62 |
ENST00000538383.1
|
NTN4
|
netrin 4 |
| chr2_+_177001685 | 1.61 |
ENST00000432796.2
|
HOXD3
|
homeobox D3 |
| chr10_-_21806759 | 1.60 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr5_-_36301984 | 1.60 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr2_+_211421262 | 1.59 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr13_-_46679185 | 1.54 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr11_-_126810521 | 1.53 |
ENST00000530572.1
|
RP11-688I9.4
|
RP11-688I9.4 |
| chr2_-_166930131 | 1.51 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr8_-_18666360 | 1.49 |
ENST00000286485.8
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr17_-_39684550 | 1.47 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chrX_+_65382433 | 1.47 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr9_-_73736511 | 1.45 |
ENST00000377110.3
ENST00000377111.2 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr17_-_77924627 | 1.45 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chrX_+_65384052 | 1.43 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr13_-_46679144 | 1.42 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr12_+_54410664 | 1.41 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr7_+_35756186 | 1.41 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr2_+_176987088 | 1.40 |
ENST00000249499.6
|
HOXD9
|
homeobox D9 |
| chrX_+_65382381 | 1.40 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr7_-_93519471 | 1.38 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chrX_+_107683096 | 1.34 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chrX_+_65384182 | 1.33 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr1_-_12679171 | 1.30 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr15_-_53097139 | 1.29 |
ENST00000560818.1
|
RP11-209K10.2
|
RP11-209K10.2 |
| chr6_+_152011628 | 1.27 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr1_+_26485511 | 1.26 |
ENST00000374268.3
|
FAM110D
|
family with sequence similarity 110, member D |
| chr15_+_69854027 | 1.25 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr7_+_35756066 | 1.25 |
ENST00000449644.1
|
AC018647.3
|
AC018647.3 |
| chr7_-_41742697 | 1.22 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chrX_-_105282712 | 1.20 |
ENST00000372563.1
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr4_-_25865159 | 1.19 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr14_-_57277163 | 1.18 |
ENST00000555006.1
|
OTX2
|
orthodenticle homeobox 2 |
| chr5_-_115872142 | 1.18 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr2_-_158295915 | 1.15 |
ENST00000418920.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr14_-_64846033 | 1.13 |
ENST00000556556.1
|
CTD-2555O16.1
|
CTD-2555O16.1 |
| chr3_-_79816965 | 1.12 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr20_+_42984330 | 1.10 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr11_-_5537920 | 1.10 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr2_+_177015122 | 1.09 |
ENST00000468418.3
|
HOXD3
|
homeobox D3 |
| chr7_-_81399438 | 1.08 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr10_-_106240032 | 1.05 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr2_-_157198860 | 1.04 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr8_-_73793975 | 1.04 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr7_+_128784712 | 1.02 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr17_+_7533439 | 1.01 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr2_-_200320768 | 1.01 |
ENST00000428695.1
|
SATB2
|
SATB homeobox 2 |
| chr9_-_124976185 | 1.01 |
ENST00000464484.2
|
LHX6
|
LIM homeobox 6 |
| chr14_-_57277178 | 1.00 |
ENST00000339475.5
ENST00000554559.1 ENST00000555804.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr7_-_75401513 | 0.98 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr7_-_81399355 | 0.98 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr18_-_77276057 | 0.94 |
ENST00000597412.1
|
AC018445.1
|
Uncharacterized protein |
| chr9_+_130026756 | 0.93 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr2_-_50574856 | 0.93 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr7_-_11871815 | 0.92 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr4_-_48683188 | 0.90 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr3_-_20053741 | 0.90 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr17_-_46690839 | 0.89 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr5_-_169626104 | 0.87 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chr19_-_19626351 | 0.86 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr8_+_38831683 | 0.85 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr12_+_70219052 | 0.85 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chrX_-_107682702 | 0.85 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr3_+_190105909 | 0.83 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr9_+_97766409 | 0.82 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr12_+_25205155 | 0.78 |
ENST00000550945.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr10_-_73497581 | 0.77 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr2_-_51259292 | 0.75 |
ENST00000401669.2
|
NRXN1
|
neurexin 1 |
| chr2_-_51259229 | 0.75 |
ENST00000405472.3
|
NRXN1
|
neurexin 1 |
| chr14_-_69444957 | 0.74 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr17_-_46657473 | 0.74 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr11_-_132812987 | 0.73 |
ENST00000541867.1
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr18_+_72922710 | 0.71 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr1_+_183774240 | 0.69 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr19_+_55996316 | 0.68 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr15_+_49715293 | 0.68 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr9_+_97766469 | 0.67 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr11_+_113185778 | 0.67 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr10_+_26505179 | 0.64 |
ENST00000376261.3
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr19_+_35634146 | 0.64 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr20_+_4702548 | 0.64 |
ENST00000305817.2
|
PRND
|
prion protein 2 (dublet) |
| chr22_-_30642782 | 0.64 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr9_-_124976154 | 0.63 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr8_+_70378852 | 0.63 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chr9_-_16705069 | 0.63 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr12_-_95510743 | 0.62 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr12_+_25205568 | 0.61 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr14_-_101295407 | 0.61 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr18_-_48351743 | 0.61 |
ENST00000588444.1
ENST00000256425.2 ENST00000428869.2 |
MRO
|
maestro |
| chr2_-_40739501 | 0.60 |
ENST00000403092.1
ENST00000402441.1 ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr7_-_81399329 | 0.60 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_+_10000640 | 0.59 |
ENST00000451755.1
|
AC006373.1
|
AC006373.1 |
| chr6_+_6588902 | 0.58 |
ENST00000230568.4
|
LY86
|
lymphocyte antigen 86 |
| chr2_+_10223908 | 0.58 |
ENST00000425235.1
|
AC104794.4
|
AC104794.4 |
| chr20_+_58713514 | 0.58 |
ENST00000432910.1
|
RP5-1043L13.1
|
RP5-1043L13.1 |
| chr11_+_34663913 | 0.58 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr7_+_141418962 | 0.57 |
ENST00000493845.1
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr9_+_105757590 | 0.57 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr1_+_183774285 | 0.57 |
ENST00000539189.1
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr11_+_124481361 | 0.56 |
ENST00000284288.2
|
PANX3
|
pannexin 3 |
| chr11_-_132813627 | 0.56 |
ENST00000374778.4
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr11_-_132813566 | 0.51 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr6_+_26156551 | 0.51 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr15_-_37783009 | 0.50 |
ENST00000561054.1
|
RP11-720L8.1
|
RP11-720L8.1 |
| chr12_-_24102576 | 0.50 |
ENST00000537393.1
ENST00000309359.1 ENST00000381381.2 ENST00000451604.2 |
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr7_-_15014398 | 0.49 |
ENST00000437998.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr10_-_24770632 | 0.48 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr6_-_64029879 | 0.48 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr11_+_34664014 | 0.48 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr3_-_51813009 | 0.48 |
ENST00000398780.3
|
IQCF6
|
IQ motif containing F6 |
| chr12_+_64173583 | 0.47 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr5_+_159656437 | 0.47 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr2_-_74726710 | 0.47 |
ENST00000377566.4
|
LBX2
|
ladybird homeobox 2 |
| chr16_-_67978016 | 0.47 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr7_-_81399411 | 0.47 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr17_+_1173853 | 0.46 |
ENST00000391429.1
|
BHLHA9
|
basic helix-loop-helix family, member a9 |
| chr16_-_21663919 | 0.46 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr3_+_173116225 | 0.46 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr9_+_44868935 | 0.46 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr4_+_114214125 | 0.46 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr3_+_164924769 | 0.45 |
ENST00000494915.1
|
RP11-85M11.2
|
RP11-85M11.2 |
| chr22_+_39916558 | 0.45 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr15_+_49715449 | 0.45 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr17_+_7461580 | 0.44 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_+_66999268 | 0.44 |
ENST00000371039.1
ENST00000424320.1 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr8_-_114449112 | 0.44 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr9_-_124989804 | 0.44 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr9_+_75229616 | 0.43 |
ENST00000340019.3
|
TMC1
|
transmembrane channel-like 1 |
| chr12_+_25205666 | 0.43 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr9_-_124990680 | 0.41 |
ENST00000541397.2
ENST00000560485.1 |
LHX6
|
LIM homeobox 6 |
| chr3_+_63428982 | 0.41 |
ENST00000479198.1
ENST00000460711.1 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr16_-_21863541 | 0.40 |
ENST00000543654.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr17_+_7461613 | 0.39 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_+_78143006 | 0.39 |
ENST00000443419.1
|
AC073628.1
|
AC073628.1 |
| chr18_-_73967160 | 0.37 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr8_-_57123815 | 0.37 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr11_-_31531121 | 0.36 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr9_-_133814455 | 0.35 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr2_+_3800169 | 0.34 |
ENST00000399143.3
|
DCDC2C
|
doublecortin domain containing 2C |
| chr12_+_130646999 | 0.34 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr11_-_27723158 | 0.34 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr3_-_45837959 | 0.30 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr3_+_41236325 | 0.30 |
ENST00000426215.1
ENST00000405570.1 |
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr11_-_84844167 | 0.30 |
ENST00000527088.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr14_+_62164340 | 0.29 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr10_-_115614127 | 0.25 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr8_+_56074008 | 0.25 |
ENST00000522559.1
|
RP11-386G21.2
|
Uncharacterized protein |
| chrX_-_100604184 | 0.24 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr8_+_26435359 | 0.24 |
ENST00000311151.5
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr3_-_45838011 | 0.23 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr18_+_55024383 | 0.23 |
ENST00000586360.1
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr5_+_176731572 | 0.23 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr12_-_71148413 | 0.22 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr7_-_81399287 | 0.21 |
ENST00000354224.6
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_122840015 | 0.21 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.9 | 3.4 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.8 | 3.9 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.8 | 9.9 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.7 | 3.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.7 | 2.2 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.7 | 2.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.6 | 3.8 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.5 | 1.6 | GO:1903718 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.5 | 3.4 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.5 | 1.8 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.4 | 2.0 | GO:0061032 | cardiac right ventricle formation(GO:0003219) visceral serous pericardium development(GO:0061032) |
| 0.4 | 1.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.4 | 2.7 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.4 | 1.1 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 2.8 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.3 | 2.4 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 | 2.4 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.3 | 1.0 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.2 | 2.7 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 1.1 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.2 | 1.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 1.8 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 0.6 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.2 | 1.8 | GO:0071638 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 | 2.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 2.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 5.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 0.5 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 2.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 4.0 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.5 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.1 | 0.3 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 1.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 1.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 1.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 1.5 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 0.5 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.7 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 2.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.6 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.1 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0007403 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 | 0.3 | GO:0051541 | neural fold elevation formation(GO:0021502) elastin metabolic process(GO:0051541) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.8 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.2 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.6 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 1.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 1.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.2 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 1.5 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.2 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.6 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.6 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 1.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.5 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.5 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 1.0 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.8 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.3 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.8 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 2.3 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 1.5 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 1.4 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 3.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.6 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 2.8 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 1.8 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 1.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 4.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 2.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 2.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 0.6 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.2 | 1.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 9.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.5 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 1.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 1.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 2.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 2.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 3.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 6.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 6.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 3.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.7 | 2.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.7 | 3.4 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.5 | 5.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 1.7 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.4 | 3.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 2.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 2.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 0.6 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 1.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.0 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.2 | 2.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 6.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 3.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 4.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.5 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 3.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.0 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 2.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.6 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.6 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.5 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.5 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 7.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 3.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 7.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 1.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 11.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 2.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 2.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 3.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 5.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 2.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 6.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 7.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 5.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 3.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 4.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 6.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 4.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 2.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 2.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |