Project
Illumina Body Map 2
Navigation
Downloads
Results for HOXC9
Z-value: 1.35
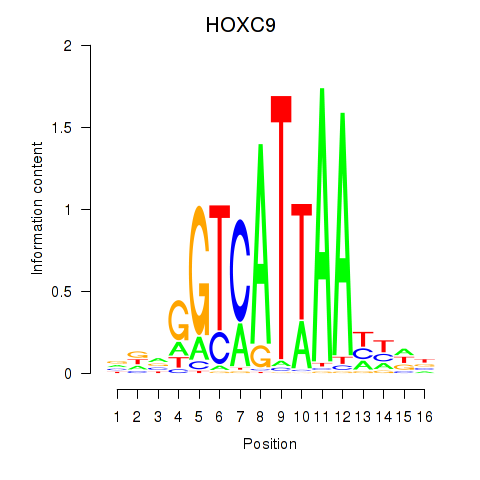
Transcription factors associated with HOXC9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC9
|
ENSG00000180806.4 | homeobox C9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC9 | hg19_v2_chr12_+_54393880_54393962 | -0.41 | 2.0e-02 | Click! |
Activity profile of HOXC9 motif
Sorted Z-values of HOXC9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_72649763 | 4.86 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr4_-_70626314 | 4.58 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr4_+_155484155 | 4.42 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr4_+_155484103 | 4.27 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr9_-_104198042 | 3.72 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr10_-_96829246 | 3.53 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr5_-_135290651 | 3.41 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr5_-_135290705 | 3.27 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr10_+_5238793 | 3.02 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr19_-_58864848 | 2.70 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr1_+_47603109 | 2.67 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr10_-_13344341 | 2.59 |
ENST00000396920.3
|
PHYH
|
phytanoyl-CoA 2-hydroxylase |
| chr12_+_20963647 | 2.54 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr5_+_36606700 | 2.41 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr15_-_90222610 | 2.37 |
ENST00000300055.5
|
PLIN1
|
perilipin 1 |
| chr15_-_90222642 | 2.35 |
ENST00000430628.2
|
PLIN1
|
perilipin 1 |
| chr17_-_9694614 | 2.35 |
ENST00000330255.5
ENST00000571134.1 |
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chr22_+_21133469 | 2.33 |
ENST00000406799.1
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr10_-_113943447 | 2.16 |
ENST00000369425.1
ENST00000348367.4 ENST00000423155.1 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr2_-_105030466 | 2.06 |
ENST00000449772.1
|
AC068535.3
|
AC068535.3 |
| chr6_+_111408698 | 2.06 |
ENST00000368851.5
|
SLC16A10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr17_+_67498295 | 2.05 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr4_+_69681710 | 2.05 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chrX_+_105936982 | 2.02 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr7_+_138915102 | 2.02 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chrX_-_15332665 | 1.99 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chrX_+_105937068 | 1.99 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr4_-_100065419 | 1.92 |
ENST00000504125.1
ENST00000505590.1 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr4_-_100065440 | 1.86 |
ENST00000508393.1
ENST00000265512.7 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr3_+_149191723 | 1.84 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr7_+_137761167 | 1.82 |
ENST00000432161.1
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr10_-_27529486 | 1.78 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr5_-_35991535 | 1.75 |
ENST00000507113.1
ENST00000274278.3 |
UGT3A1
|
UDP glycosyltransferase 3 family, polypeptide A1 |
| chr4_-_69536346 | 1.74 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr4_-_100065389 | 1.73 |
ENST00000512499.1
|
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr2_-_169887827 | 1.66 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr14_+_60712463 | 1.65 |
ENST00000325642.3
ENST00000529574.1 |
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr12_-_21757774 | 1.65 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr16_+_72090053 | 1.63 |
ENST00000576168.2
ENST00000567185.3 ENST00000567612.2 |
HP
|
haptoglobin |
| chr17_+_67498538 | 1.58 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr8_-_81083890 | 1.55 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr8_-_17555164 | 1.54 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr14_+_74083548 | 1.52 |
ENST00000381139.1
|
ACOT6
|
acyl-CoA thioesterase 6 |
| chr2_-_200715834 | 1.50 |
ENST00000420128.1
ENST00000416668.1 |
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr12_+_20963632 | 1.49 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chrM_+_10464 | 1.48 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr11_+_22694123 | 1.47 |
ENST00000534801.1
|
GAS2
|
growth arrest-specific 2 |
| chr18_-_74839891 | 1.42 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr3_-_196242233 | 1.41 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr8_-_81083731 | 1.41 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr16_-_48281305 | 1.41 |
ENST00000356608.2
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr17_-_9683238 | 1.38 |
ENST00000571771.1
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chrX_+_43515467 | 1.36 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr16_-_48281444 | 1.33 |
ENST00000537808.1
ENST00000569991.1 |
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr1_+_220863187 | 1.32 |
ENST00000294889.5
|
C1orf115
|
chromosome 1 open reading frame 115 |
| chr7_-_44580861 | 1.31 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr7_-_113559104 | 1.30 |
ENST00000284601.3
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A |
| chr12_+_104235229 | 1.30 |
ENST00000551650.1
|
RP11-650K20.3
|
Uncharacterized protein |
| chr19_-_36304201 | 1.30 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr2_-_230096756 | 1.29 |
ENST00000354069.6
|
PID1
|
phosphotyrosine interaction domain containing 1 |
| chr17_+_7348658 | 1.28 |
ENST00000570557.1
ENST00000536404.2 ENST00000576360.1 |
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr6_+_143447322 | 1.28 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr4_-_70626430 | 1.26 |
ENST00000310613.3
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr8_-_42396185 | 1.25 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr4_+_187148556 | 1.22 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr7_+_5468362 | 1.20 |
ENST00000608012.1
|
RP11-1275H24.3
|
RP11-1275H24.3 |
| chr10_-_115614127 | 1.19 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr1_+_202385953 | 1.16 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr2_+_27719697 | 1.14 |
ENST00000264717.2
ENST00000424318.2 |
GCKR
|
glucokinase (hexokinase 4) regulator |
| chr8_-_105479270 | 1.11 |
ENST00000521573.2
ENST00000351513.2 |
DPYS
|
dihydropyrimidinase |
| chr14_+_39944025 | 1.09 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr2_-_200715573 | 1.09 |
ENST00000420922.2
|
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr14_+_67291158 | 1.08 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr9_-_95055923 | 1.07 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr14_+_21387491 | 1.06 |
ENST00000258817.2
|
RP11-84C10.2
|
RP11-84C10.2 |
| chr11_+_19798964 | 1.06 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr8_-_17579726 | 1.06 |
ENST00000381861.3
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr16_+_71560154 | 1.04 |
ENST00000539698.3
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr8_+_145215928 | 1.02 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr16_-_12897642 | 1.02 |
ENST00000433677.2
ENST00000261660.4 ENST00000381774.4 |
CPPED1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr16_-_66584059 | 1.02 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr11_+_65266507 | 1.00 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr9_-_93405352 | 0.99 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr14_+_21359558 | 0.98 |
ENST00000304639.3
|
RNASE3
|
ribonuclease, RNase A family, 3 |
| chr8_-_82608409 | 0.98 |
ENST00000518568.1
|
SLC10A5
|
solute carrier family 10, member 5 |
| chr4_+_70894130 | 0.98 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr6_+_158733692 | 0.98 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr11_+_46366918 | 0.97 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr8_+_9009202 | 0.97 |
ENST00000518496.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr7_-_111424462 | 0.97 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr2_+_234526272 | 0.96 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr8_-_80993010 | 0.95 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr17_-_28257080 | 0.94 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr16_+_71560023 | 0.94 |
ENST00000572450.1
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr10_-_94257512 | 0.94 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr8_+_104033296 | 0.94 |
ENST00000521514.1
ENST00000518738.1 |
ATP6V1C1
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 |
| chr11_+_22688150 | 0.93 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr5_-_65018834 | 0.93 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr17_+_58018269 | 0.93 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr12_+_25348186 | 0.93 |
ENST00000555711.1
ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5
|
LYR motif containing 5 |
| chr16_-_66907139 | 0.92 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr7_-_111424506 | 0.92 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr8_+_104033277 | 0.91 |
ENST00000518857.1
ENST00000395862.3 |
ATP6V1C1
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 |
| chr1_+_212965170 | 0.91 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr1_+_12524965 | 0.90 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr12_+_25348139 | 0.90 |
ENST00000557540.2
ENST00000381356.4 |
LYRM5
|
LYR motif containing 5 |
| chr1_-_212004090 | 0.88 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr1_+_241695424 | 0.88 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr6_+_131571535 | 0.87 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr16_-_66583994 | 0.87 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr12_+_58176525 | 0.87 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr20_-_45980621 | 0.86 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr18_+_32558380 | 0.86 |
ENST00000588349.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr10_+_24755416 | 0.85 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr4_+_71600063 | 0.84 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr13_-_52027134 | 0.84 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr11_-_13517565 | 0.83 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr1_+_40862501 | 0.82 |
ENST00000539317.1
|
SMAP2
|
small ArfGAP2 |
| chr20_+_57414743 | 0.81 |
ENST00000313949.7
|
GNAS
|
GNAS complex locus |
| chr9_+_80912059 | 0.81 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr5_-_148442584 | 0.80 |
ENST00000394358.2
ENST00000512049.1 |
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chrX_+_120181457 | 0.80 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr10_+_115614370 | 0.79 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr20_-_36152914 | 0.79 |
ENST00000397131.1
|
BLCAP
|
bladder cancer associated protein |
| chr9_-_21187598 | 0.77 |
ENST00000421715.1
|
IFNA4
|
interferon, alpha 4 |
| chr12_-_118628315 | 0.77 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr6_-_135424186 | 0.77 |
ENST00000529882.1
|
HBS1L
|
HBS1-like (S. cerevisiae) |
| chrX_+_150565038 | 0.77 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr3_+_16306691 | 0.76 |
ENST00000285083.5
ENST00000605932.1 ENST00000435829.2 |
OXNAD1
|
oxidoreductase NAD-binding domain containing 1 |
| chr20_+_5987890 | 0.75 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr1_-_19426149 | 0.74 |
ENST00000429347.2
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr7_+_80275663 | 0.74 |
ENST00000413265.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_-_54355430 | 0.74 |
ENST00000371399.1
ENST00000072644.1 ENST00000412288.1 |
YIPF1
|
Yip1 domain family, member 1 |
| chr20_+_57414795 | 0.73 |
ENST00000371098.2
ENST00000371075.3 |
GNAS
|
GNAS complex locus |
| chr9_-_130889990 | 0.72 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr7_+_7196565 | 0.71 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr16_+_58074069 | 0.71 |
ENST00000570065.1
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr10_-_127505167 | 0.71 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chrM_+_10053 | 0.70 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr11_+_22688615 | 0.70 |
ENST00000533363.1
|
GAS2
|
growth arrest-specific 2 |
| chrX_+_139791917 | 0.70 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr12_-_63328817 | 0.70 |
ENST00000228705.6
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr8_+_132916318 | 0.69 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr2_-_122262593 | 0.69 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr12_-_118628350 | 0.69 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr2_-_172750733 | 0.67 |
ENST00000392592.4
ENST00000422440.2 |
SLC25A12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr2_+_48541776 | 0.66 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr10_-_24770632 | 0.66 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr17_-_42767092 | 0.66 |
ENST00000588687.1
|
CCDC43
|
coiled-coil domain containing 43 |
| chr14_+_53173910 | 0.66 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr17_+_55162453 | 0.65 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr1_+_146714291 | 0.65 |
ENST00000431239.1
ENST00000369259.3 ENST00000369258.4 ENST00000361293.5 |
CHD1L
|
chromodomain helicase DNA binding protein 1-like |
| chr16_-_66583701 | 0.65 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr6_-_52859968 | 0.64 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr6_-_109702885 | 0.64 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr2_+_233390890 | 0.63 |
ENST00000258385.3
ENST00000536614.1 ENST00000457943.2 |
CHRND
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr17_-_10372875 | 0.63 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr14_+_97925151 | 0.63 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr12_-_90049878 | 0.63 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr20_+_30697298 | 0.62 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr8_-_62602327 | 0.62 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr2_+_233390863 | 0.61 |
ENST00000449596.1
ENST00000543200.1 |
CHRND
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr4_-_85654615 | 0.61 |
ENST00000514711.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr4_-_69111401 | 0.61 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr9_+_103947311 | 0.60 |
ENST00000395056.2
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr21_-_27423339 | 0.60 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr8_-_93978333 | 0.59 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr12_-_90049828 | 0.59 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr2_+_62132781 | 0.59 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr2_+_207630081 | 0.58 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr14_+_21387508 | 0.58 |
ENST00000555624.1
|
RP11-84C10.2
|
RP11-84C10.2 |
| chr14_+_53173890 | 0.58 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr9_-_95055956 | 0.58 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr6_-_146057144 | 0.58 |
ENST00000367519.3
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr5_-_139930713 | 0.57 |
ENST00000602657.1
|
SRA1
|
steroid receptor RNA activator 1 |
| chr18_+_32558208 | 0.57 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr17_+_44803922 | 0.57 |
ENST00000465370.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr9_-_95056010 | 0.57 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr2_-_200320768 | 0.57 |
ENST00000428695.1
|
SATB2
|
SATB homeobox 2 |
| chr2_+_187371440 | 0.57 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr8_-_93978216 | 0.56 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr8_-_93978346 | 0.56 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr7_+_80275621 | 0.56 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr8_-_93978357 | 0.56 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr16_+_53412368 | 0.56 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr9_+_131549610 | 0.54 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr1_-_113478603 | 0.54 |
ENST00000443580.1
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr12_+_32687221 | 0.54 |
ENST00000525053.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr9_-_69262509 | 0.54 |
ENST00000377449.1
ENST00000382399.4 ENST00000377439.1 ENST00000377441.1 ENST00000377457.5 |
CBWD6
|
COBW domain containing 6 |
| chr1_-_212965104 | 0.54 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr12_-_11091862 | 0.54 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr2_+_62132800 | 0.53 |
ENST00000538736.1
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr10_-_128110441 | 0.53 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr6_-_83903600 | 0.52 |
ENST00000506587.1
ENST00000507554.1 |
PGM3
|
phosphoglucomutase 3 |
| chr9_-_70490107 | 0.52 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr4_-_176733377 | 0.52 |
ENST00000505375.1
|
GPM6A
|
glycoprotein M6A |
| chr1_-_205091115 | 0.51 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr18_-_12656715 | 0.51 |
ENST00000462226.1
ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1
|
spire-type actin nucleation factor 1 |
| chr3_+_69811858 | 0.51 |
ENST00000433517.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr8_-_93978309 | 0.50 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.8 | 3.9 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.7 | 9.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.6 | 3.7 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.6 | 2.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.5 | 1.6 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.5 | 2.6 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.5 | 2.5 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.4 | 1.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.4 | 3.0 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.4 | 1.8 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.4 | 1.1 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.3 | 2.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 5.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 4.0 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.3 | 0.8 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 1.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.3 | 1.1 | GO:0019482 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.3 | 3.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.3 | 1.3 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 2.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 1.4 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.2 | 1.9 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 0.7 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.2 | 0.9 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.2 | 0.9 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 6.8 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.2 | 1.7 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.2 | 1.8 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 1.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 1.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 1.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 2.2 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 0.5 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.2 | 0.7 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 1.5 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 0.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 2.1 | GO:0070327 | aromatic amino acid transport(GO:0015801) thyroid hormone transport(GO:0070327) |
| 0.2 | 0.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 2.0 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.2 | 4.9 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 1.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 1.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 1.4 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 1.2 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.8 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.1 | 0.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.5 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.6 | GO:0071873 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.5 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 1.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.6 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.5 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 | 0.3 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 1.5 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 2.7 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.1 | 0.4 | GO:1904501 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 | 1.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 1.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 4.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 0.9 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.8 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.3 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.5 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 1.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.4 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 1.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.6 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.6 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.5 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.3 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 1.9 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 3.8 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.2 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.3 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 1.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.9 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.3 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 1.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.4 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.2 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.7 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 1.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.4 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 1.0 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.6 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 2.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.4 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.3 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.6 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.3 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.5 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.3 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 4.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 2.9 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 0.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.4 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.2 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.2 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.4 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 2.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0072513 | semicircular canal morphogenesis(GO:0048752) positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.3 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 1.8 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 1.0 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.9 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.1 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 1.2 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.8 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 3.5 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0046426 | regulation of JAK-STAT cascade(GO:0046425) negative regulation of JAK-STAT cascade(GO:0046426) regulation of STAT cascade(GO:1904892) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.9 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.6 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.4 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 4.5 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.2 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.6 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.8 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.7 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 4.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 1.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 1.9 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 1.7 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.2 | 0.7 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.2 | 0.6 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.1 | 1.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 1.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 2.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 3.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.9 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 0.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.6 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.6 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 4.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0070369 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 3.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 4.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.7 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 1.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 3.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 1.1 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 1.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.6 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 5.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.6 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.3 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 2.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 4.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0019115 | benzaldehyde dehydrogenase activity(GO:0019115) |
| 1.6 | 4.9 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.9 | 2.6 | GO:0048244 | phytanoyl-CoA dioxygenase activity(GO:0048244) |
| 0.7 | 2.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.7 | 3.5 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.6 | 1.8 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.6 | 3.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.6 | 2.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.6 | 1.7 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.5 | 2.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.4 | 2.5 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.4 | 1.1 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.3 | 1.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.3 | 0.9 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.3 | 3.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 0.9 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.3 | 1.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 0.8 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.2 | 2.0 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 2.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 1.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 0.9 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.2 | 2.7 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 1.1 | GO:0002054 | nucleobase binding(GO:0002054) dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 5.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 0.6 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 2.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 0.8 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 0.5 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.2 | 5.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 0.7 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.2 | 1.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.7 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.2 | 1.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.7 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 3.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 6.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.5 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 2.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 3.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 1.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.4 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.9 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 0.4 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 1.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.5 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 1.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.2 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 1.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 9.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 1.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 1.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 1.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 2.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 1.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 3.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 4.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 8.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 4.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 4.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 4.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 3.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.7 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 5.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 4.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 5.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 3.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 4.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 4.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 4.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 4.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 3.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 3.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 4.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 4.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 4.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 2.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 2.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |