Project
Illumina Body Map 2
Navigation
Downloads
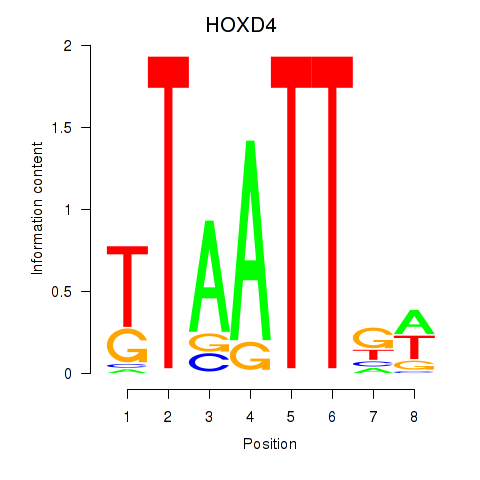
Results for HOXD4
Z-value: 0.91
Transcription factors associated with HOXD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD4
|
ENSG00000170166.5 | homeobox D4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD4 | hg19_v2_chr2_+_177015950_177015950 | -0.12 | 5.0e-01 | Click! |
Activity profile of HOXD4 motif
Sorted Z-values of HOXD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_48483736 | 2.64 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr4_+_155484155 | 2.60 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr4_+_155484103 | 2.55 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr5_+_101569696 | 2.08 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr15_+_84115868 | 1.83 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr6_-_87804815 | 1.64 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr10_-_50970322 | 1.46 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr2_+_155555201 | 1.45 |
ENST00000544049.1
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr3_-_170744498 | 1.45 |
ENST00000382808.4
ENST00000314251.3 |
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr20_-_45980621 | 1.26 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr3_-_190167571 | 1.25 |
ENST00000354905.2
|
TMEM207
|
transmembrane protein 207 |
| chr4_-_36245561 | 1.21 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr4_-_70505358 | 1.20 |
ENST00000457664.2
ENST00000604629.1 ENST00000604021.1 |
UGT2A2
|
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chr8_+_50822344 | 1.17 |
ENST00000518864.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr3_+_142342228 | 1.17 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr2_+_234590556 | 1.12 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr6_+_123317116 | 1.11 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr7_+_123488124 | 1.09 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chrX_+_37850026 | 0.98 |
ENST00000341016.3
|
CXorf27
|
chromosome X open reading frame 27 |
| chr15_-_54025300 | 0.96 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr4_+_74347400 | 0.92 |
ENST00000226355.3
|
AFM
|
afamin |
| chr17_-_50235423 | 0.92 |
ENST00000340813.6
|
CA10
|
carbonic anhydrase X |
| chr10_-_98119030 | 0.92 |
ENST00000371172.3
ENST00000393871.1 ENST00000419479.1 |
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr2_+_191045656 | 0.89 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr1_+_197237352 | 0.87 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr12_-_39734783 | 0.87 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr20_-_43729750 | 0.86 |
ENST00000537075.1
ENST00000306117.1 |
KCNS1
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
| chr17_-_64225508 | 0.83 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr10_-_61899124 | 0.81 |
ENST00000373815.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr8_+_79503458 | 0.79 |
ENST00000518467.1
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr1_-_190446759 | 0.79 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr2_-_166060552 | 0.78 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr7_+_29186174 | 0.75 |
ENST00000439384.1
|
CHN2
|
chimerin 2 |
| chr5_-_150727111 | 0.72 |
ENST00000335244.4
ENST00000521967.1 |
SLC36A2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr1_-_238108575 | 0.71 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr2_-_31361543 | 0.71 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr18_-_74839891 | 0.68 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr5_-_13944652 | 0.68 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr4_+_146539415 | 0.68 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr2_+_166095898 | 0.68 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr2_-_159237472 | 0.67 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr2_-_165424973 | 0.67 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr9_-_93405352 | 0.66 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr2_-_163695128 | 0.65 |
ENST00000332142.5
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr2_+_171034646 | 0.65 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr5_-_78809950 | 0.64 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chrX_+_37865804 | 0.64 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr18_+_22040620 | 0.63 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr18_+_68002675 | 0.63 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chrX_+_149867681 | 0.63 |
ENST00000438018.1
ENST00000436701.1 |
MTMR1
|
myotubularin related protein 1 |
| chr3_+_111718173 | 0.60 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr2_-_225434538 | 0.60 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chrX_+_56590002 | 0.59 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr8_-_40200877 | 0.58 |
ENST00000521030.1
|
CTA-392C11.1
|
CTA-392C11.1 |
| chr18_-_61311485 | 0.57 |
ENST00000436264.1
ENST00000356424.6 ENST00000341074.5 |
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| chrX_+_130192318 | 0.56 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr5_-_55412774 | 0.56 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr4_+_69313145 | 0.54 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr1_+_149754227 | 0.53 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr1_+_115572415 | 0.53 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr15_-_94614049 | 0.53 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr7_+_29186192 | 0.52 |
ENST00000539406.1
|
CHN2
|
chimerin 2 |
| chr2_-_166060571 | 0.51 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr3_+_142342240 | 0.50 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr7_-_15014398 | 0.50 |
ENST00000437998.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr3_+_158787098 | 0.50 |
ENST00000397832.2
ENST00000451172.1 ENST00000482126.1 |
IQCJ
|
IQ motif containing J |
| chr8_+_42552503 | 0.50 |
ENST00000534391.1
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3 (neuronal) |
| chr13_-_61989655 | 0.49 |
ENST00000409204.4
|
PCDH20
|
protocadherin 20 |
| chr4_-_46996424 | 0.49 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr1_-_151762900 | 0.48 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr1_-_182921119 | 0.47 |
ENST00000423786.1
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like |
| chr17_-_78450398 | 0.46 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr1_-_28384598 | 0.45 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr1_+_42846443 | 0.44 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chr15_+_54793169 | 0.44 |
ENST00000559093.1
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr2_-_163695238 | 0.44 |
ENST00000328032.4
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr12_-_81763184 | 0.43 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr5_-_110848253 | 0.43 |
ENST00000505803.1
ENST00000502322.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr10_-_17171785 | 0.43 |
ENST00000377823.1
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr20_-_29978286 | 0.42 |
ENST00000376315.2
|
DEFB119
|
defensin, beta 119 |
| chr8_-_27941359 | 0.42 |
ENST00000418860.1
|
NUGGC
|
nuclear GTPase, germinal center associated |
| chr4_-_76944621 | 0.42 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr1_-_151762943 | 0.42 |
ENST00000368825.3
ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH
|
tudor and KH domain containing |
| chr3_+_178276488 | 0.42 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr1_-_120935894 | 0.41 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr13_-_45768841 | 0.40 |
ENST00000379108.1
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr14_+_29236269 | 0.39 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr8_-_42698292 | 0.39 |
ENST00000529779.1
|
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr3_-_18480173 | 0.39 |
ENST00000414509.1
|
SATB1
|
SATB homeobox 1 |
| chr2_+_234826016 | 0.38 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr6_-_136847099 | 0.38 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr7_+_101460882 | 0.38 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr9_+_97562440 | 0.37 |
ENST00000395357.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr16_-_3350614 | 0.37 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr4_+_146560245 | 0.36 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr16_+_84801852 | 0.36 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr7_-_38293030 | 0.36 |
ENST00000390333.1
|
TRGJ2
|
T cell receptor gamma joining 2 |
| chr9_-_7961080 | 0.35 |
ENST00000435444.1
|
RP11-29B9.2
|
RP11-29B9.2 |
| chr6_-_49931818 | 0.35 |
ENST00000322066.3
|
DEFB114
|
defensin, beta 114 |
| chr11_-_36310958 | 0.35 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr11_-_57519253 | 0.35 |
ENST00000422652.1
ENST00000527995.1 |
BTBD18
|
BTB (POZ) domain containing 18 |
| chr2_-_99279928 | 0.35 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr4_-_89442940 | 0.34 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr11_-_16419067 | 0.34 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr15_-_64386120 | 0.34 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr7_-_121944491 | 0.33 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr18_+_32402321 | 0.32 |
ENST00000587723.1
|
DTNA
|
dystrobrevin, alpha |
| chr10_+_69869237 | 0.32 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr15_-_64385981 | 0.32 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr3_-_157221380 | 0.31 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr16_-_29910853 | 0.31 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr11_-_55371874 | 0.31 |
ENST00000302231.4
|
OR4C11
|
olfactory receptor, family 4, subfamily C, member 11 |
| chr1_+_84630574 | 0.30 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_-_118628315 | 0.30 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr1_-_231560790 | 0.30 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr3_-_18480260 | 0.30 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr1_+_109265033 | 0.30 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr12_+_32654965 | 0.29 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr14_-_99947121 | 0.29 |
ENST00000329331.3
ENST00000436070.2 |
SETD3
|
SET domain containing 3 |
| chr6_+_72926145 | 0.29 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr16_+_87636474 | 0.28 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr1_+_214161854 | 0.28 |
ENST00000435016.1
|
PROX1
|
prospero homeobox 1 |
| chr13_+_37581115 | 0.28 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr2_+_224822121 | 0.28 |
ENST00000258383.3
|
MRPL44
|
mitochondrial ribosomal protein L44 |
| chr5_-_160279207 | 0.27 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr2_+_88047606 | 0.27 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr3_-_178103080 | 0.27 |
ENST00000414475.1
|
RP11-33A14.1
|
RP11-33A14.1 |
| chr7_-_137028498 | 0.27 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr1_+_182419261 | 0.27 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr8_-_72274467 | 0.27 |
ENST00000340726.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr11_-_72492878 | 0.26 |
ENST00000535054.1
ENST00000545082.1 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr4_+_130014836 | 0.26 |
ENST00000502887.1
|
C4orf33
|
chromosome 4 open reading frame 33 |
| chr12_+_28605426 | 0.26 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr14_-_99947168 | 0.26 |
ENST00000331768.5
|
SETD3
|
SET domain containing 3 |
| chr17_+_41132564 | 0.26 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chrX_-_92928557 | 0.25 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr12_+_9980113 | 0.25 |
ENST00000537723.1
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr9_+_273038 | 0.25 |
ENST00000487230.1
ENST00000469391.1 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr2_-_207024233 | 0.25 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr7_+_77428149 | 0.25 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr12_+_16109519 | 0.25 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr15_-_56209306 | 0.25 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr2_-_235363231 | 0.24 |
ENST00000439798.1
|
AC097713.4
|
AC097713.4 |
| chr7_+_55433131 | 0.24 |
ENST00000254770.2
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr3_+_97483572 | 0.24 |
ENST00000335979.2
ENST00000394206.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr7_-_122339162 | 0.24 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr3_+_140981456 | 0.24 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr12_+_96196875 | 0.24 |
ENST00000553095.1
|
RP11-536G4.1
|
Uncharacterized protein |
| chr2_+_204571375 | 0.23 |
ENST00000374478.4
|
CD28
|
CD28 molecule |
| chr7_+_77428066 | 0.23 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr18_-_19283649 | 0.23 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr1_+_84630645 | 0.23 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_+_158787041 | 0.23 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr14_-_21058982 | 0.23 |
ENST00000556526.1
|
RNASE12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr11_+_48510269 | 0.22 |
ENST00000446524.1
|
OR4A47
|
olfactory receptor, family 4, subfamily A, member 47 |
| chr6_+_132873832 | 0.22 |
ENST00000275200.1
|
TAAR8
|
trace amine associated receptor 8 |
| chr5_-_110848189 | 0.22 |
ENST00000296632.3
ENST00000512160.1 ENST00000509887.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr8_-_42698433 | 0.22 |
ENST00000345117.2
ENST00000254250.3 |
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr7_-_77427676 | 0.22 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr8_-_42396185 | 0.21 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr4_-_153601136 | 0.21 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr11_+_112041253 | 0.21 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chr5_-_54988448 | 0.21 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr4_-_130014609 | 0.21 |
ENST00000511426.1
|
SCLT1
|
sodium channel and clathrin linker 1 |
| chr18_-_2982869 | 0.21 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr4_-_130014705 | 0.20 |
ENST00000503401.1
|
SCLT1
|
sodium channel and clathrin linker 1 |
| chr2_+_27886330 | 0.20 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr20_+_32150140 | 0.20 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr2_+_32502952 | 0.20 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr18_-_19994830 | 0.20 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr14_+_23067146 | 0.20 |
ENST00000428304.2
|
ABHD4
|
abhydrolase domain containing 4 |
| chr3_+_51575596 | 0.20 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr3_-_186524144 | 0.20 |
ENST00000427785.1
|
RFC4
|
replication factor C (activator 1) 4, 37kDa |
| chr15_+_55700741 | 0.20 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr1_-_74949033 | 0.19 |
ENST00000416014.2
|
LRRC53
|
leucine rich repeat containing 53 |
| chr5_+_59783941 | 0.19 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr6_+_142468361 | 0.19 |
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr8_-_57026541 | 0.19 |
ENST00000311923.1
|
MOS
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr1_+_109256067 | 0.19 |
ENST00000271311.2
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr15_+_36887069 | 0.19 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr5_-_137475071 | 0.18 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr13_+_73629107 | 0.18 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chrX_-_13835147 | 0.18 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr1_+_67632083 | 0.18 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr18_+_32556892 | 0.17 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr17_+_9479944 | 0.17 |
ENST00000396219.3
ENST00000352665.5 |
WDR16
|
WD repeat domain 16 |
| chr10_+_95848824 | 0.17 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr7_+_63361201 | 0.17 |
ENST00000450544.1
|
RP11-340I6.8
|
RP11-340I6.8 |
| chr14_+_20611895 | 0.17 |
ENST00000333629.1
|
OR4N5
|
olfactory receptor, family 4, subfamily N, member 5 |
| chr4_+_146601356 | 0.17 |
ENST00000438731.1
ENST00000511965.1 |
C4orf51
|
chromosome 4 open reading frame 51 |
| chr21_+_35736302 | 0.16 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr6_+_49467671 | 0.16 |
ENST00000371197.4
ENST00000545705.1 |
GLYATL3
|
glycine-N-acyltransferase-like 3 |
| chr14_+_23025534 | 0.16 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr1_+_170115142 | 0.16 |
ENST00000439373.2
|
METTL11B
|
methyltransferase like 11B |
| chr6_-_15548591 | 0.16 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr9_-_107361788 | 0.16 |
ENST00000374779.2
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5 |
| chr12_+_21168630 | 0.15 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr18_-_69449517 | 0.15 |
ENST00000584810.1
|
RP11-723G8.2
|
Uncharacterized protein |
| chr17_+_3118915 | 0.15 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr7_-_104909435 | 0.15 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 1.5 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.3 | 1.0 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.2 | 1.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.7 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 0.2 | 0.6 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 0.6 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 1.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.2 | 1.3 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 | 0.6 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.8 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.4 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 1.5 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 0.6 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.7 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.3 | GO:0090425 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.3 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.3 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.1 | 0.7 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.8 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.2 | GO:1902102 | metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 0.2 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.4 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.3 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.5 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 1.0 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 2.4 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.2 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 1.0 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 2.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.1 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 1.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.3 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.0 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 2.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905024) regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905031) |
| 0.0 | 0.2 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 1.1 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.5 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.9 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 1.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 2.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.9 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 2.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 3.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.3 | 1.5 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 0.9 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 1.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 1.5 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.1 | 0.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 1.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.7 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 2.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.3 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 5.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.0 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 1.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 1.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.7 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 1.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 2.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 1.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 3.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |