Project
Illumina Body Map 2
Navigation
Downloads
Results for IKZF2
Z-value: 1.52
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.12 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | hg19_v2_chr2_-_214014959_214015058 | 0.36 | 4.4e-02 | Click! |
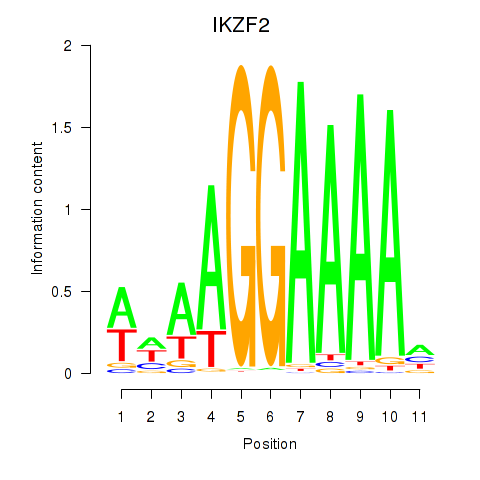
Activity profile of IKZF2 motif
Sorted Z-values of IKZF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_71532207 | 6.36 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr4_-_71532601 | 6.34 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr4_-_71532339 | 5.68 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr14_-_106552755 | 4.17 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr4_+_68424434 | 4.02 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr5_+_35852797 | 3.51 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr12_-_122240792 | 3.43 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr14_-_106926724 | 3.35 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr14_-_106668095 | 3.25 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr14_-_107114267 | 3.18 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr12_-_68845165 | 3.01 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr15_-_20193370 | 2.85 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr3_-_161089289 | 2.82 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr17_-_46262541 | 2.68 |
ENST00000579336.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr2_+_98330009 | 2.65 |
ENST00000264972.5
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa |
| chr1_+_171217622 | 2.48 |
ENST00000433267.1
ENST00000367750.3 |
FMO1
|
flavin containing monooxygenase 1 |
| chr14_-_106610852 | 2.47 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr5_-_34043310 | 2.38 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr1_+_171217677 | 2.37 |
ENST00000402921.2
|
FMO1
|
flavin containing monooxygenase 1 |
| chr4_-_109088940 | 2.35 |
ENST00000438313.2
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr12_-_68845417 | 2.28 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr16_-_33647696 | 2.18 |
ENST00000558425.1
ENST00000569103.2 |
RP11-812E19.9
|
Uncharacterized protein |
| chr22_+_40297079 | 2.17 |
ENST00000344138.4
ENST00000543252.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr11_+_58938903 | 2.15 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr1_-_47655686 | 2.14 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr21_-_34185944 | 2.09 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chrX_+_135730297 | 2.06 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr5_-_111091948 | 2.01 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr1_+_101185290 | 1.99 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr2_+_114163945 | 1.97 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr2_-_190044480 | 1.91 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chrX_+_135730373 | 1.88 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr7_-_142139783 | 1.87 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6 |
| chrX_-_45629661 | 1.87 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr1_+_209941942 | 1.84 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr11_+_71903169 | 1.79 |
ENST00000393676.3
|
FOLR1
|
folate receptor 1 (adult) |
| chr14_+_91709279 | 1.78 |
ENST00000554096.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr7_-_142099977 | 1.73 |
ENST00000390359.3
|
TRBV7-8
|
T cell receptor beta variable 7-8 |
| chr1_+_9711781 | 1.72 |
ENST00000536656.1
ENST00000377346.4 |
PIK3CD
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr14_+_91709103 | 1.70 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr7_-_16505440 | 1.69 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr14_+_64932210 | 1.65 |
ENST00000394718.4
|
AKAP5
|
A kinase (PRKA) anchor protein 5 |
| chr2_+_7865923 | 1.64 |
ENST00000417930.1
|
AC092580.4
|
AC092580.4 |
| chr3_-_183273477 | 1.62 |
ENST00000341319.3
|
KLHL6
|
kelch-like family member 6 |
| chr1_-_206306107 | 1.57 |
ENST00000436158.1
ENST00000455672.1 |
RP11-38J22.6
|
RP11-38J22.6 |
| chr3_+_238273 | 1.54 |
ENST00000256509.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr20_+_58179582 | 1.53 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr2_+_204801471 | 1.51 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr7_-_127672146 | 1.49 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr17_-_46688334 | 1.48 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr4_-_114900831 | 1.47 |
ENST00000315366.7
|
ARSJ
|
arylsulfatase family, member J |
| chr6_-_31846744 | 1.46 |
ENST00000414427.1
ENST00000229729.6 ENST00000375562.4 |
SLC44A4
|
solute carrier family 44, member 4 |
| chr3_+_177159744 | 1.45 |
ENST00000439009.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr2_+_177028805 | 1.45 |
ENST00000249440.3
|
HOXD3
|
homeobox D3 |
| chr14_+_22748980 | 1.43 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr8_+_80523321 | 1.42 |
ENST00000518111.1
|
STMN2
|
stathmin-like 2 |
| chr3_+_46395219 | 1.42 |
ENST00000445132.2
ENST00000292301.4 |
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr12_+_116985896 | 1.41 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr21_+_26934165 | 1.40 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr11_-_104893863 | 1.40 |
ENST00000260315.3
ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5
|
caspase 5, apoptosis-related cysteine peptidase |
| chr6_-_159466136 | 1.40 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr17_+_7792101 | 1.39 |
ENST00000358181.4
ENST00000330494.7 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr5_+_140180635 | 1.38 |
ENST00000522353.2
ENST00000532566.2 |
PCDHA3
|
protocadherin alpha 3 |
| chr9_-_73483926 | 1.38 |
ENST00000396283.1
ENST00000361823.5 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr12_-_6580094 | 1.36 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr5_+_92919043 | 1.36 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr3_-_27764190 | 1.35 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr3_-_112329110 | 1.34 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr13_-_30160925 | 1.32 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr2_-_85637459 | 1.31 |
ENST00000409921.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr4_-_87278857 | 1.31 |
ENST00000509464.1
ENST00000511167.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr7_-_127671674 | 1.30 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr12_+_93130311 | 1.30 |
ENST00000344636.3
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr6_+_27107053 | 1.29 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr11_-_75017734 | 1.29 |
ENST00000532525.1
|
ARRB1
|
arrestin, beta 1 |
| chr11_+_61522844 | 1.29 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr12_+_50690489 | 1.27 |
ENST00000598429.1
|
AC140061.12
|
Uncharacterized protein |
| chr14_-_60337684 | 1.25 |
ENST00000267484.5
|
RTN1
|
reticulon 1 |
| chr3_+_177159695 | 1.24 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr6_-_159466042 | 1.24 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr15_-_55541227 | 1.23 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr6_-_41715128 | 1.21 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr6_+_106546808 | 1.21 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr6_+_31638156 | 1.21 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr12_-_109025849 | 1.21 |
ENST00000228463.6
|
SELPLG
|
selectin P ligand |
| chr9_-_117692697 | 1.21 |
ENST00000223795.2
|
TNFSF8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr9_-_16705069 | 1.20 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr21_+_39628852 | 1.20 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr19_-_42947121 | 1.18 |
ENST00000601181.1
|
CXCL17
|
chemokine (C-X-C motif) ligand 17 |
| chr1_+_158801095 | 1.16 |
ENST00000368141.4
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr21_-_15918618 | 1.16 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr19_+_24009879 | 1.16 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chrX_+_73164149 | 1.14 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr17_+_35294075 | 1.14 |
ENST00000254457.5
|
LHX1
|
LIM homeobox 1 |
| chr11_-_33891362 | 1.13 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr19_-_47290535 | 1.13 |
ENST00000412532.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr15_+_81299370 | 1.12 |
ENST00000560091.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr11_-_67141090 | 1.12 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr3_+_238427 | 1.10 |
ENST00000397491.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr22_+_40297105 | 1.10 |
ENST00000540310.1
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr15_+_81475047 | 1.09 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr1_-_169599314 | 1.08 |
ENST00000367786.2
ENST00000458599.2 ENST00000367795.2 ENST00000263686.6 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr9_+_26746951 | 1.08 |
ENST00000523363.1
|
RP11-18A15.1
|
RP11-18A15.1 |
| chrX_+_78003204 | 1.08 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr9_-_3469181 | 1.07 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr6_+_167536230 | 1.07 |
ENST00000341935.5
ENST00000349984.4 |
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr8_-_23282820 | 1.06 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr17_-_48785216 | 1.06 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr11_+_122526383 | 1.06 |
ENST00000284273.5
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B |
| chr1_-_169599353 | 1.05 |
ENST00000367793.2
ENST00000367794.2 ENST00000367792.2 ENST00000367791.2 ENST00000367788.2 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr1_-_242612779 | 1.03 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr6_+_106534192 | 1.02 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr13_-_99959641 | 1.01 |
ENST00000376414.4
|
GPR183
|
G protein-coupled receptor 183 |
| chr6_+_31637944 | 1.01 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr1_+_174933899 | 1.01 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr3_-_121740969 | 1.00 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chrX_-_100662881 | 1.00 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chr6_+_147830063 | 1.00 |
ENST00000367474.1
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr4_-_123377880 | 1.00 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chr15_-_66545995 | 0.99 |
ENST00000395614.1
ENST00000288745.3 ENST00000422354.1 ENST00000395625.2 ENST00000360698.4 ENST00000409699.2 |
MEGF11
|
multiple EGF-like-domains 11 |
| chr2_+_232135245 | 0.98 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr21_+_39628780 | 0.98 |
ENST00000417042.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr9_+_132099158 | 0.98 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chrX_-_133119476 | 0.98 |
ENST00000543339.1
|
GPC3
|
glypican 3 |
| chr4_-_74486217 | 0.98 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr16_+_81272287 | 0.97 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr2_+_27301435 | 0.97 |
ENST00000380320.4
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr6_-_160679905 | 0.95 |
ENST00000366953.3
|
SLC22A2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr16_+_50313426 | 0.95 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chrX_+_123097014 | 0.95 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr3_+_118905564 | 0.95 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
| chr6_-_112081113 | 0.93 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr19_-_41859814 | 0.93 |
ENST00000221930.5
|
TGFB1
|
transforming growth factor, beta 1 |
| chr17_-_26127525 | 0.92 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chr15_-_90358048 | 0.91 |
ENST00000300060.6
ENST00000560137.1 |
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr4_+_158141899 | 0.91 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr2_-_134326009 | 0.91 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr6_-_11779403 | 0.91 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr14_-_52436247 | 0.90 |
ENST00000597846.1
|
AL358333.1
|
HCG2013195; Uncharacterized protein |
| chrX_+_73164167 | 0.90 |
ENST00000414209.1
ENST00000602895.1 ENST00000453317.1 ENST00000602546.1 ENST00000602985.1 ENST00000415215.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr4_-_74486347 | 0.90 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr8_-_82024290 | 0.90 |
ENST00000220597.4
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr6_+_33388013 | 0.88 |
ENST00000449372.2
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chr11_-_15643937 | 0.88 |
ENST00000533082.1
|
RP11-531H8.2
|
RP11-531H8.2 |
| chr2_-_237416071 | 0.88 |
ENST00000309507.5
ENST00000431676.2 |
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr1_+_111888890 | 0.87 |
ENST00000369738.4
|
PIFO
|
primary cilia formation |
| chr8_+_80523962 | 0.86 |
ENST00000518491.1
|
STMN2
|
stathmin-like 2 |
| chr3_+_46395579 | 0.85 |
ENST00000421659.1
|
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr4_+_158141843 | 0.85 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chrX_+_133507389 | 0.84 |
ENST00000370800.4
|
PHF6
|
PHD finger protein 6 |
| chr13_-_46756351 | 0.84 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr3_+_10068095 | 0.84 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr20_+_42574317 | 0.84 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr12_-_123921256 | 0.83 |
ENST00000280571.8
|
RILPL2
|
Rab interacting lysosomal protein-like 2 |
| chr6_+_21593972 | 0.80 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr12_-_50677255 | 0.80 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr11_-_67141640 | 0.80 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr3_+_178276488 | 0.80 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr1_+_158979792 | 0.79 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr6_+_106988986 | 0.78 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr4_+_158141806 | 0.78 |
ENST00000393815.2
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr20_-_3996036 | 0.78 |
ENST00000336095.6
|
RNF24
|
ring finger protein 24 |
| chr8_-_133637624 | 0.77 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr6_+_26365387 | 0.77 |
ENST00000532865.1
ENST00000530653.1 ENST00000527417.1 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr8_-_134114721 | 0.77 |
ENST00000522119.1
ENST00000523610.1 ENST00000521302.1 ENST00000519558.1 ENST00000519747.1 ENST00000517648.1 |
SLA
|
Src-like-adaptor |
| chr6_-_152639479 | 0.77 |
ENST00000356820.4
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr6_+_149539053 | 0.77 |
ENST00000451095.1
|
RP1-111D6.3
|
RP1-111D6.3 |
| chr17_-_56082455 | 0.77 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr4_-_185303418 | 0.76 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chrX_-_15683147 | 0.76 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr6_+_26365443 | 0.76 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr8_-_16043780 | 0.76 |
ENST00000445506.2
|
MSR1
|
macrophage scavenger receptor 1 |
| chr13_+_58206655 | 0.76 |
ENST00000377918.3
|
PCDH17
|
protocadherin 17 |
| chr3_+_193853927 | 0.75 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr4_+_96012585 | 0.75 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr8_+_26240414 | 0.75 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr1_+_36038971 | 0.75 |
ENST00000373235.3
|
TFAP2E
|
transcription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) |
| chr7_-_122840015 | 0.75 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr1_+_158979686 | 0.74 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr18_-_35145728 | 0.74 |
ENST00000361795.5
ENST00000603232.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr8_+_104831554 | 0.74 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr4_-_185776854 | 0.73 |
ENST00000511703.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr18_-_5540384 | 0.73 |
ENST00000584670.1
ENST00000582703.1 ENST00000580179.1 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr13_+_20268547 | 0.73 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr3_+_178525080 | 0.72 |
ENST00000358316.3
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr1_+_158979680 | 0.72 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr17_+_55055466 | 0.71 |
ENST00000262288.3
ENST00000572710.1 ENST00000575395.1 |
SCPEP1
|
serine carboxypeptidase 1 |
| chr6_+_13925318 | 0.71 |
ENST00000423553.2
ENST00000537388.1 |
RNF182
|
ring finger protein 182 |
| chr10_+_119302508 | 0.71 |
ENST00000442245.4
|
EMX2
|
empty spiracles homeobox 2 |
| chr7_-_138482933 | 0.71 |
ENST00000310018.2
|
ATP6V0A4
|
ATPase, H+ transporting, lysosomal V0 subunit a4 |
| chr2_-_158182322 | 0.70 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr4_-_11431389 | 0.70 |
ENST00000002596.5
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr20_+_15177480 | 0.70 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr7_-_138482849 | 0.70 |
ENST00000353492.4
|
ATP6V0A4
|
ATPase, H+ transporting, lysosomal V0 subunit a4 |
| chr11_+_65383227 | 0.69 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr15_-_34880646 | 0.69 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr7_-_111424506 | 0.69 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr13_+_49551020 | 0.68 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr14_+_22988947 | 0.68 |
ENST00000390513.1
|
TRAJ24
|
T cell receptor alpha joining 24 |
| chr19_+_18496957 | 0.68 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IKZF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 1.1 | 3.3 | GO:2000458 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.7 | 2.2 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.6 | 1.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.6 | 1.8 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.6 | 2.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.5 | 4.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.5 | 1.5 | GO:0048925 | lateral line system development(GO:0048925) |
| 0.5 | 1.4 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.4 | 2.6 | GO:0043366 | beta selection(GO:0043366) |
| 0.4 | 0.4 | GO:0045861 | negative regulation of proteolysis(GO:0045861) |
| 0.4 | 1.6 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.4 | 1.2 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.4 | 1.1 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.4 | 2.8 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.3 | 0.7 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.3 | 1.7 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 2.4 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.3 | 0.9 | GO:0044416 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.3 | 1.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.3 | 1.7 | GO:0060374 | positive regulation of neutrophil apoptotic process(GO:0033031) mast cell differentiation(GO:0060374) |
| 0.3 | 2.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.3 | 1.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.3 | 2.7 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 1.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.3 | 1.0 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.3 | 0.8 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.2 | 1.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 3.5 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 0.7 | GO:0043132 | NAD transport(GO:0043132) |
| 0.2 | 0.9 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 1.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.2 | 1.9 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.2 | 0.8 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 1.2 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 1.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 14.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 1.2 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 1.4 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 1.3 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 1.0 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.1 | 0.4 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.1 | 1.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.8 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.5 | GO:0002572 | pro-T cell differentiation(GO:0002572) regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 2.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.5 | GO:0090678 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 3.8 | GO:0051023 | regulation of immunoglobulin secretion(GO:0051023) |
| 0.1 | 1.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 2.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 1.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 1.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 1.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 2.7 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 1.8 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.8 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 1.4 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.7 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 1.0 | GO:0016139 | glycoside catabolic process(GO:0016139) glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 2.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 1.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 0.7 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.8 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.9 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.4 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.6 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.9 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.1 | 0.4 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.9 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.1 | 1.5 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.9 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 1.1 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.1 | 1.7 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.3 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 2.3 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 0.2 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.4 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.1 | 0.7 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.4 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 2.6 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 1.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.4 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 1.9 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.1 | 1.0 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.1 | 0.2 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 1.5 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.7 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.6 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 1.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.4 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.2 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.7 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.8 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.9 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.4 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 5.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.8 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 1.7 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 1.2 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 2.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.4 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.5 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.7 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 4.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 2.2 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.0 | 0.7 | GO:0090179 | eyelid development in camera-type eye(GO:0061029) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 1.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 1.0 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 1.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.8 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.7 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.6 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.7 | GO:0006023 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.9 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.3 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 2.6 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 4.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.4 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.8 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.3 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.2 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.3 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 1.4 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.3 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.6 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.5 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.2 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.6 | 1.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 1.9 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.4 | 2.5 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.3 | 15.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 0.7 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 2.8 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 1.4 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.2 | 1.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.2 | 2.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 1.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 5.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.4 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 2.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 1.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.4 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 1.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 1.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.7 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 1.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 8.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 5.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 1.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 1.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 2.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 2.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 2.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 1.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.8 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.9 | 3.5 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.5 | 4.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 4.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.5 | 2.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 1.8 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.4 | 1.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.4 | 3.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.4 | 2.8 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 1.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.3 | 1.0 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.3 | 1.1 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.2 | 1.0 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.2 | 0.9 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.2 | 15.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 1.9 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 1.7 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 1.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 0.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 1.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 1.5 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.2 | 1.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 0.9 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 1.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.7 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 2.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 1.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 1.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.4 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 1.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.9 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 1.1 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 2.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 2.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.7 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 1.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.6 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.7 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 0.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 7.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 3.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 1.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.5 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 1.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.4 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.7 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.5 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 2.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 2.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 1.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 1.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.3 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 2.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.9 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 1.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 4.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.5 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.5 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 1.0 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 1.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 1.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 6.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 6.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 2.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 3.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 3.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 3.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 4.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 3.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 4.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 3.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 3.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 3.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 4.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 2.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 6.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.8 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.0 | 1.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 2.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 2.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 3.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 3.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 6.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |