Project
Illumina Body Map 2
Navigation
Downloads
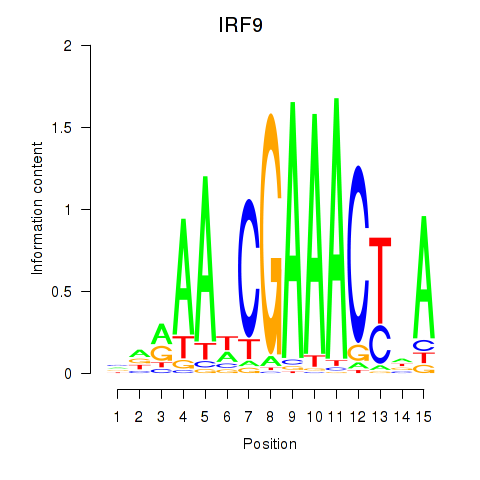
Results for IRF9
Z-value: 1.20
Transcription factors associated with IRF9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF9
|
ENSG00000213928.4 | interferon regulatory factor 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF9 | hg19_v2_chr14_+_24630465_24630531 | 0.33 | 6.3e-02 | Click! |
Activity profile of IRF9 motif
Sorted Z-values of IRF9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_161600990 | 5.93 |
ENST00000531221.1
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
| chr1_-_161600942 | 5.72 |
ENST00000421702.2
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
| chr6_+_6588902 | 5.08 |
ENST00000230568.4
|
LY86
|
lymphocyte antigen 86 |
| chr3_-_27764190 | 5.01 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr1_-_161600822 | 4.86 |
ENST00000534776.1
ENST00000540048.1 |
FCGR3B
FCGR3A
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr22_+_40297105 | 4.80 |
ENST00000540310.1
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr21_+_42733870 | 4.62 |
ENST00000330714.3
ENST00000436410.1 ENST00000435611.1 |
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr22_+_40297079 | 4.47 |
ENST00000344138.4
ENST00000543252.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr1_-_150738261 | 4.45 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr12_+_113344755 | 3.88 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr12_-_121477039 | 3.58 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr19_+_16254488 | 3.51 |
ENST00000588246.1
ENST00000593031.1 |
HSH2D
|
hematopoietic SH2 domain containing |
| chr12_-_121476959 | 3.38 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr12_+_113344582 | 3.37 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr4_-_76944621 | 3.37 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr21_+_42797958 | 3.33 |
ENST00000419044.1
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr12_-_121476750 | 3.31 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr18_-_67624160 | 3.24 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr6_-_24936170 | 3.20 |
ENST00000538035.1
|
FAM65B
|
family with sequence similarity 65, member B |
| chr21_+_42798158 | 3.19 |
ENST00000441677.1
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chrX_+_37639302 | 2.95 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr2_-_7005785 | 2.83 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr6_+_106546808 | 2.78 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr2_+_7005959 | 2.73 |
ENST00000442639.1
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr21_+_42798124 | 2.71 |
ENST00000417963.1
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr18_-_67623906 | 2.68 |
ENST00000583955.1
|
CD226
|
CD226 molecule |
| chr12_+_113344811 | 2.64 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr21_+_42798094 | 2.63 |
ENST00000398598.3
ENST00000455164.2 ENST00000424365.1 |
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr18_-_67624412 | 2.59 |
ENST00000580335.1
|
CD226
|
CD226 molecule |
| chr12_+_113376157 | 2.35 |
ENST00000228928.7
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr14_-_67981916 | 2.31 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chrX_+_37639264 | 2.31 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr1_+_66820058 | 2.29 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr8_+_27238147 | 2.26 |
ENST00000412793.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr1_+_241695670 | 2.23 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr6_-_31324943 | 2.22 |
ENST00000412585.2
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr1_+_241695424 | 2.20 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr6_-_33281979 | 2.17 |
ENST00000426633.2
ENST00000467025.1 |
TAPBP
|
TAP binding protein (tapasin) |
| chr17_-_54991369 | 2.13 |
ENST00000537230.1
|
TRIM25
|
tripartite motif containing 25 |
| chr20_-_56195449 | 2.12 |
ENST00000541799.1
|
ZBP1
|
Z-DNA binding protein 1 |
| chr20_-_56195525 | 2.09 |
ENST00000371173.3
ENST00000395822.3 ENST00000340462.4 ENST00000343535.4 |
ZBP1
|
Z-DNA binding protein 1 |
| chr13_-_46961580 | 2.07 |
ENST00000378787.3
ENST00000378797.2 ENST00000429979.1 ENST00000378781.3 |
KIAA0226L
|
KIAA0226-like |
| chr4_-_169401628 | 2.03 |
ENST00000514748.1
ENST00000512371.1 ENST00000260184.7 ENST00000505890.1 ENST00000511577.1 |
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
| chr3_+_122283175 | 2.02 |
ENST00000383661.3
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr6_-_33282163 | 2.01 |
ENST00000434618.2
ENST00000456592.2 |
TAPBP
|
TAP binding protein (tapasin) |
| chr6_-_33282024 | 2.00 |
ENST00000475304.1
ENST00000489157.1 |
TAPBP
|
TAP binding protein (tapasin) |
| chr1_+_79086088 | 1.99 |
ENST00000370751.5
ENST00000342282.3 |
IFI44L
|
interferon-induced protein 44-like |
| chr14_-_67981870 | 1.94 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr16_-_74734672 | 1.89 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr17_+_6659153 | 1.89 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr15_+_45003675 | 1.88 |
ENST00000558401.1
ENST00000559916.1 ENST00000544417.1 |
B2M
|
beta-2-microglobulin |
| chr2_+_163175394 | 1.83 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr7_+_18535893 | 1.80 |
ENST00000432645.2
ENST00000441542.2 |
HDAC9
|
histone deacetylase 9 |
| chr2_+_7017796 | 1.65 |
ENST00000382040.3
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr17_+_78234625 | 1.60 |
ENST00000508628.2
ENST00000582970.1 ENST00000456466.1 ENST00000319921.4 |
RNF213
|
ring finger protein 213 |
| chr11_-_615570 | 1.57 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr17_+_6658878 | 1.57 |
ENST00000574394.1
|
XAF1
|
XIAP associated factor 1 |
| chr2_-_152146385 | 1.57 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr11_-_57334732 | 1.51 |
ENST00000526659.1
ENST00000527022.1 |
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr6_-_32806483 | 1.50 |
ENST00000374899.4
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr3_-_142166796 | 1.50 |
ENST00000392981.2
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr3_+_122399697 | 1.48 |
ENST00000494811.1
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr7_+_134832808 | 1.42 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr8_-_27941359 | 1.41 |
ENST00000418860.1
|
NUGGC
|
nuclear GTPase, germinal center associated |
| chr13_-_43566301 | 1.40 |
ENST00000398762.3
ENST00000313640.7 ENST00000313624.7 |
EPSTI1
|
epithelial stromal interaction 1 (breast) |
| chr12_+_6881678 | 1.37 |
ENST00000441671.2
ENST00000203629.2 |
LAG3
|
lymphocyte-activation gene 3 |
| chr4_-_147442817 | 1.33 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr18_+_13382553 | 1.33 |
ENST00000586222.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr22_+_42148515 | 1.30 |
ENST00000540880.1
|
MEI1
|
meiosis inhibitor 1 |
| chr3_+_187086120 | 1.28 |
ENST00000259030.2
|
RTP4
|
receptor (chemosensory) transporter protein 4 |
| chr15_-_26108355 | 1.24 |
ENST00000356865.6
|
ATP10A
|
ATPase, class V, type 10A |
| chr1_+_79115503 | 1.24 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr17_+_41363854 | 1.23 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr14_+_100531738 | 1.23 |
ENST00000555706.1
|
EVL
|
Enah/Vasp-like |
| chr10_-_91174215 | 1.22 |
ENST00000371837.1
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase |
| chr1_+_948803 | 1.21 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr16_-_28518153 | 1.21 |
ENST00000356897.1
|
IL27
|
interleukin 27 |
| chr3_-_130745403 | 1.20 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr3_+_122283064 | 1.18 |
ENST00000296161.4
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr3_-_142166904 | 1.18 |
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr11_-_4414880 | 1.15 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr3_-_121379739 | 1.14 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr9_-_32526184 | 1.14 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr10_+_91061712 | 1.11 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr6_-_32806506 | 1.11 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr17_-_54991395 | 1.11 |
ENST00000316881.4
|
TRIM25
|
tripartite motif containing 25 |
| chr3_-_122283079 | 1.10 |
ENST00000471785.1
ENST00000466126.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_+_151986709 | 1.06 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr11_-_60720002 | 1.04 |
ENST00000538739.1
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr3_-_122283100 | 1.03 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr11_-_67141090 | 1.03 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr2_+_33701707 | 1.02 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chrX_-_39956656 | 0.99 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr19_+_17516494 | 0.97 |
ENST00000534306.1
|
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr11_-_615942 | 0.96 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr11_-_57335280 | 0.96 |
ENST00000287156.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr12_-_56753858 | 0.96 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr1_+_158979792 | 0.94 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr17_-_74489215 | 0.92 |
ENST00000585701.1
ENST00000591192.1 ENST00000589526.1 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr11_+_5646213 | 0.90 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr10_+_91174486 | 0.89 |
ENST00000416601.1
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr3_-_146262352 | 0.88 |
ENST00000462666.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr12_+_113376249 | 0.86 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr17_+_7211656 | 0.84 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr3_-_146262365 | 0.84 |
ENST00000448787.2
|
PLSCR1
|
phospholipid scramblase 1 |
| chr1_-_154580616 | 0.83 |
ENST00000368474.4
|
ADAR
|
adenosine deaminase, RNA-specific |
| chr3_+_122399444 | 0.82 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr6_-_82462425 | 0.81 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr14_+_24630465 | 0.80 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr4_-_147442982 | 0.78 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr1_+_158979680 | 0.74 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr18_+_3262954 | 0.72 |
ENST00000584539.1
|
MYL12B
|
myosin, light chain 12B, regulatory |
| chr1_+_154377669 | 0.71 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr10_-_95242044 | 0.71 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr3_-_146262637 | 0.70 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr3_-_142166846 | 0.69 |
ENST00000463916.1
ENST00000544157.1 |
XRN1
|
5'-3' exoribonuclease 1 |
| chr1_+_158979686 | 0.69 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr16_-_15736881 | 0.69 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr3_-_179169181 | 0.68 |
ENST00000497513.1
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr3_-_122283424 | 0.67 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr10_-_95241951 | 0.64 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr3_-_28389922 | 0.60 |
ENST00000415852.1
|
AZI2
|
5-azacytidine induced 2 |
| chr10_+_91174314 | 0.58 |
ENST00000371795.4
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr19_+_10196981 | 0.58 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr7_-_105332084 | 0.56 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr3_-_146262488 | 0.56 |
ENST00000487389.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr15_+_58702742 | 0.54 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr2_+_7073174 | 0.54 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr3_-_146262428 | 0.52 |
ENST00000486631.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr16_+_53412368 | 0.51 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr5_-_142783694 | 0.51 |
ENST00000394466.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr3_-_146262293 | 0.48 |
ENST00000448205.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr5_-_142784003 | 0.43 |
ENST00000416954.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr4_+_37892682 | 0.43 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr20_+_61436146 | 0.42 |
ENST00000290291.6
|
OGFR
|
opioid growth factor receptor |
| chr19_+_17516624 | 0.38 |
ENST00000596322.1
ENST00000600008.1 ENST00000601885.1 |
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr13_+_50070077 | 0.37 |
ENST00000378319.3
ENST00000426879.1 |
PHF11
|
PHD finger protein 11 |
| chr14_-_57277163 | 0.34 |
ENST00000555006.1
|
OTX2
|
orthodenticle homeobox 2 |
| chr5_-_95297678 | 0.33 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr2_+_33701684 | 0.32 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr12_-_27167233 | 0.32 |
ENST00000535819.1
ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3
|
transmembrane 7 superfamily member 3 |
| chr20_+_11898507 | 0.31 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr13_+_50070491 | 0.30 |
ENST00000496612.1
ENST00000357596.3 ENST00000485919.1 ENST00000442195.1 |
PHF11
|
PHD finger protein 11 |
| chr2_-_106810742 | 0.29 |
ENST00000409501.3
ENST00000428048.2 ENST00000441952.1 ENST00000457835.1 ENST00000540130.1 |
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chrX_+_22056165 | 0.26 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr2_-_106810783 | 0.25 |
ENST00000283148.7
|
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr6_+_37400974 | 0.22 |
ENST00000455891.1
ENST00000373451.4 |
CMTR1
|
cap methyltransferase 1 |
| chr4_-_169239921 | 0.22 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr19_+_10222189 | 0.21 |
ENST00000321826.4
|
P2RY11
|
purinergic receptor P2Y, G-protein coupled, 11 |
| chr1_-_182558374 | 0.21 |
ENST00000367559.3
ENST00000539397.1 |
RNASEL
|
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent) |
| chr17_+_38296576 | 0.18 |
ENST00000264645.7
|
CASC3
|
cancer susceptibility candidate 3 |
| chr2_-_152118276 | 0.18 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr17_-_4167142 | 0.17 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr6_+_126240442 | 0.12 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr9_+_102668915 | 0.11 |
ENST00000259400.6
ENST00000531035.1 ENST00000525640.1 ENST00000534052.1 ENST00000526607.1 |
STX17
|
syntaxin 17 |
| chr3_-_46037299 | 0.11 |
ENST00000296137.2
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chrX_-_63005405 | 0.09 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr3_-_141747459 | 0.08 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr17_-_40264692 | 0.04 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr5_-_137911049 | 0.03 |
ENST00000297185.3
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin) |
| chr5_-_95297534 | 0.02 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr2_-_55920952 | 0.01 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr14_+_100531615 | 0.01 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chr1_+_236687881 | 0.01 |
ENST00000526634.1
|
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0050823 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 1.7 | 5.0 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.4 | 8.5 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 1.1 | 4.5 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 1.1 | 5.3 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.9 | 2.8 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.7 | 16.5 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.7 | 4.4 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.7 | 2.8 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.7 | 3.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.6 | 4.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.5 | 5.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.4 | 2.5 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.4 | 4.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.4 | 2.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.3 | 2.3 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.3 | 1.6 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 3.5 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.3 | 1.2 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.3 | 28.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.3 | 2.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.3 | 3.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) negative regulation of myoblast fusion(GO:1901740) |
| 0.2 | 1.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.2 | 1.4 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.2 | 1.6 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.2 | 5.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 0.5 | GO:0033320 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.2 | 2.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 0.7 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 2.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 16.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 3.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.0 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.1 | 1.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 2.5 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.1 | 1.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 1.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 1.3 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 1.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 9.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 3.2 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.1 | 1.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.3 | GO:0045141 | meiotic telomere clustering(GO:0045141) |
| 0.0 | 1.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.3 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 1.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 3.9 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.4 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 1.2 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.9 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 1.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.6 | 4.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.4 | 1.9 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 5.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 2.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 0.7 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.1 | 0.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 2.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 16.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 9.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 4.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 6.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 4.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 10.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 4.4 | GO:0031968 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 1.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 4.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 16.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 23.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 1.8 | 8.8 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 1.0 | 5.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.9 | 16.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.7 | 2.8 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.7 | 3.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.4 | 3.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 2.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 9.7 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.3 | 1.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 0.7 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.2 | 2.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 1.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 0.5 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.2 | 1.0 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.2 | 1.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 4.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 1.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 3.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 2.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 4.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 12.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 4.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 2.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.8 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 2.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 2.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 16.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 2.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 6.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 4.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 3.2 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 5.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 6.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 2.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 5.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 3.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 3.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 4.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 47.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 2.9 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 10.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 3.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 3.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 13.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 5.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 3.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 2.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 3.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 5.1 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 2.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 2.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |