Project
Illumina Body Map 2
Navigation
Downloads
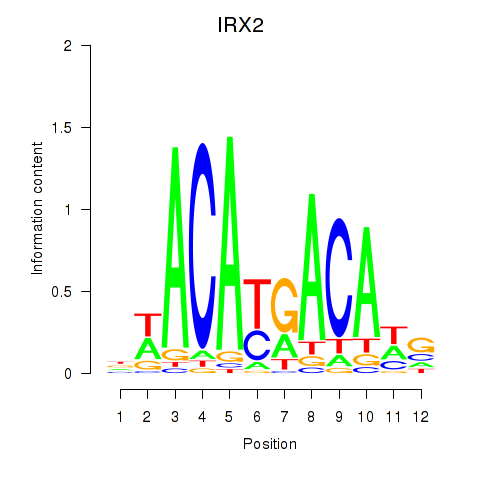
Results for IRX2
Z-value: 1.29
Transcription factors associated with IRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX2
|
ENSG00000170561.8 | iroquois homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX2 | hg19_v2_chr5_-_2751762_2751784 | 0.14 | 4.5e-01 | Click! |
Activity profile of IRX2 motif
Sorted Z-values of IRX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_61122220 | 6.39 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr2_+_168675182 | 3.63 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr4_+_88896819 | 3.62 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr5_+_36608422 | 3.59 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr1_+_166958346 | 3.50 |
ENST00000367872.4
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr1_+_166958497 | 3.48 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr21_-_22175341 | 2.95 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr1_+_166958504 | 2.87 |
ENST00000447624.1
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr19_-_51466681 | 2.82 |
ENST00000456750.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr16_+_58537737 | 2.80 |
ENST00000561738.1
|
NDRG4
|
NDRG family member 4 |
| chr21_-_22175450 | 2.77 |
ENST00000435279.2
|
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chrX_-_132095419 | 2.70 |
ENST00000370836.2
ENST00000521489.1 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr15_-_44486632 | 2.66 |
ENST00000484674.1
|
FRMD5
|
FERM domain containing 5 |
| chr4_-_168155417 | 2.65 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_+_169575875 | 2.60 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr10_-_23528745 | 2.58 |
ENST00000376501.5
|
C10orf115
|
chromosome 10 open reading frame 115 |
| chr2_+_171640291 | 2.56 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr4_+_158141843 | 2.47 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr4_+_158142750 | 2.30 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr14_+_29236269 | 2.20 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr10_-_61122625 | 2.19 |
ENST00000468840.2
|
FAM13C
|
family with sequence similarity 13, member C |
| chr8_-_82359662 | 2.19 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr18_+_50278430 | 2.18 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chrX_-_138790348 | 2.17 |
ENST00000414978.1
ENST00000519895.1 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr18_+_55018044 | 2.15 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr15_+_38226827 | 2.09 |
ENST00000559502.1
ENST00000558148.1 ENST00000558158.1 |
TMCO5A
|
transmembrane and coiled-coil domains 5A |
| chr4_+_114214125 | 2.06 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr1_-_190446759 | 2.03 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr14_+_62462541 | 2.00 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr11_+_98891797 | 2.00 |
ENST00000527185.1
ENST00000528682.1 ENST00000524871.1 |
CNTN5
|
contactin 5 |
| chr9_-_79267432 | 1.97 |
ENST00000424866.1
|
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr9_-_73736511 | 1.94 |
ENST00000377110.3
ENST00000377111.2 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr2_+_48844937 | 1.93 |
ENST00000448460.1
ENST00000437125.1 ENST00000430487.2 |
GTF2A1L
|
general transcription factor IIA, 1-like |
| chr3_+_169539710 | 1.89 |
ENST00000340806.6
|
LRRIQ4
|
leucine-rich repeats and IQ motif containing 4 |
| chr18_+_32455201 | 1.86 |
ENST00000590831.2
|
DTNA
|
dystrobrevin, alpha |
| chr9_-_23779367 | 1.84 |
ENST00000440102.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr19_-_55953704 | 1.83 |
ENST00000416792.1
|
SHISA7
|
shisa family member 7 |
| chr9_+_34957477 | 1.80 |
ENST00000544237.1
|
KIAA1045
|
KIAA1045 |
| chrX_+_150869023 | 1.79 |
ENST00000448324.1
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr4_+_158141899 | 1.79 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr6_+_88117683 | 1.78 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr16_-_20367584 | 1.77 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr17_-_26127525 | 1.70 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chr14_+_75536280 | 1.69 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr19_-_38720294 | 1.68 |
ENST00000412732.1
ENST00000456296.1 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr10_-_61122582 | 1.67 |
ENST00000503444.1
|
FAM13C
|
family with sequence similarity 13, member C |
| chr5_+_161277603 | 1.65 |
ENST00000519621.1
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr19_-_35719609 | 1.64 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187, member B |
| chr14_+_50999744 | 1.63 |
ENST00000441560.2
|
ATL1
|
atlastin GTPase 1 |
| chr7_-_123673471 | 1.62 |
ENST00000455783.1
|
TMEM229A
|
transmembrane protein 229A |
| chr5_-_138210977 | 1.60 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chrX_-_100129128 | 1.60 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr14_+_32964258 | 1.59 |
ENST00000556638.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr11_+_33037652 | 1.58 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr2_-_233877912 | 1.57 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr9_+_12695702 | 1.56 |
ENST00000381136.2
|
TYRP1
|
tyrosinase-related protein 1 |
| chr1_-_238649319 | 1.53 |
ENST00000400946.2
|
RP11-371I1.2
|
long intergenic non-protein coding RNA 1139 |
| chrX_-_117119243 | 1.51 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr11_-_62477313 | 1.50 |
ENST00000464544.1
ENST00000530009.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr4_+_57371509 | 1.49 |
ENST00000360096.2
|
ARL9
|
ADP-ribosylation factor-like 9 |
| chr12_-_16758059 | 1.49 |
ENST00000261169.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_+_10292308 | 1.48 |
ENST00000377081.1
|
KIF1B
|
kinesin family member 1B |
| chrX_+_148855726 | 1.47 |
ENST00000370416.4
|
HSFX1
|
heat shock transcription factor family, X linked 1 |
| chr3_+_94657016 | 1.41 |
ENST00000462219.1
|
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr2_+_48844973 | 1.40 |
ENST00000403751.3
|
GTF2A1L
|
general transcription factor IIA, 1-like |
| chr8_-_124665190 | 1.40 |
ENST00000325995.7
|
KLHL38
|
kelch-like family member 38 |
| chrX_+_30261847 | 1.39 |
ENST00000378981.3
ENST00000397550.1 |
MAGEB1
|
melanoma antigen family B, 1 |
| chr4_+_159131596 | 1.39 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr7_-_14880892 | 1.36 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr12_-_16758304 | 1.35 |
ENST00000320122.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_-_173572181 | 1.35 |
ENST00000536496.1
ENST00000367714.3 |
SLC9C2
|
solute carrier family 9, member C2 (putative) |
| chr8_+_67405794 | 1.34 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chrX_-_148676974 | 1.34 |
ENST00000524178.1
|
HSFX2
|
heat shock transcription factor family, X linked 2 |
| chr10_+_127661942 | 1.33 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr1_+_204839959 | 1.30 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr6_-_50016364 | 1.28 |
ENST00000322246.4
|
DEFB112
|
defensin, beta 112 |
| chr1_+_108918421 | 1.27 |
ENST00000444143.2
ENST00000294652.8 |
NBPF6
|
neuroblastoma breakpoint family, member 6 |
| chr19_-_38720354 | 1.25 |
ENST00000416611.1
|
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chrX_+_107020963 | 1.25 |
ENST00000509000.2
|
NCBP2L
|
nuclear cap binding protein subunit 2-like |
| chr4_-_80247162 | 1.24 |
ENST00000286794.4
|
NAA11
|
N(alpha)-acetyltransferase 11, NatA catalytic subunit |
| chr22_-_36013368 | 1.23 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr1_+_113009163 | 1.23 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr9_+_35792151 | 1.22 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr4_+_83821835 | 1.22 |
ENST00000302236.5
|
THAP9
|
THAP domain containing 9 |
| chr7_-_7603957 | 1.21 |
ENST00000608807.1
|
RP5-1159O4.1
|
RP5-1159O4.1 |
| chr5_-_138211051 | 1.20 |
ENST00000518785.1
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr6_+_46661575 | 1.19 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr9_-_113100088 | 1.18 |
ENST00000374510.4
ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa) |
| chr14_+_75536335 | 1.17 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr14_-_65289812 | 1.17 |
ENST00000389720.3
ENST00000389721.5 ENST00000389722.3 |
SPTB
|
spectrin, beta, erythrocytic |
| chr16_+_81272287 | 1.17 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr12_-_91572278 | 1.13 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr19_+_13134772 | 1.12 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr18_+_5748793 | 1.11 |
ENST00000566533.1
ENST00000562452.2 |
RP11-945C19.1
|
RP11-945C19.1 |
| chr16_-_32687431 | 1.10 |
ENST00000398680.3
ENST00000569420.1 |
TP53TG3
|
TP53 target 3 |
| chr11_+_103907308 | 1.10 |
ENST00000302259.3
|
DDI1
|
DNA-damage inducible 1 homolog 1 (S. cerevisiae) |
| chr4_+_159131630 | 1.10 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr3_+_132843652 | 1.07 |
ENST00000508711.1
|
TMEM108
|
transmembrane protein 108 |
| chr1_+_29138654 | 1.05 |
ENST00000234961.2
|
OPRD1
|
opioid receptor, delta 1 |
| chrX_-_73061339 | 1.03 |
ENST00000602863.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr11_+_124789146 | 1.03 |
ENST00000408930.5
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1 |
| chr3_+_259218 | 1.03 |
ENST00000449294.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr11_+_111126707 | 1.02 |
ENST00000280325.4
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chrX_-_74742846 | 1.01 |
ENST00000373361.3
|
ZDHHC15
|
zinc finger, DHHC-type containing 15 |
| chr13_+_35516390 | 1.00 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr14_+_101295638 | 1.00 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr20_-_31592113 | 0.99 |
ENST00000420875.1
ENST00000375519.2 ENST00000375523.3 ENST00000356173.3 |
SUN5
|
Sad1 and UNC84 domain containing 5 |
| chr1_-_151148442 | 0.97 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr15_-_45670924 | 0.94 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr10_-_28270795 | 0.94 |
ENST00000545014.1
|
ARMC4
|
armadillo repeat containing 4 |
| chr12_+_112856690 | 0.93 |
ENST00000392597.1
ENST00000351677.2 |
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr14_+_101295948 | 0.93 |
ENST00000452514.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr12_+_80730292 | 0.92 |
ENST00000298820.3
|
OTOGL
|
otogelin-like |
| chr1_+_16083154 | 0.92 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr16_+_33205585 | 0.92 |
ENST00000360260.2
ENST00000398666.3 |
TP53TG3C
|
TP53 target 3C |
| chr16_+_33262120 | 0.92 |
ENST00000569741.1
ENST00000380147.3 |
TP53TG3B
|
TP53 target 3B |
| chr1_-_151148492 | 0.91 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr10_+_134150835 | 0.90 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr20_-_29978286 | 0.90 |
ENST00000376315.2
|
DEFB119
|
defensin, beta 119 |
| chr16_-_12062333 | 0.89 |
ENST00000597717.1
|
AC007216.2
|
Uncharacterized protein |
| chrX_-_100129320 | 0.89 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr3_+_183815318 | 0.88 |
ENST00000425359.2
|
HTR3E
|
5-hydroxytryptamine (serotonin) receptor 3E, ionotropic |
| chr1_-_109399682 | 0.87 |
ENST00000369995.3
ENST00000370001.3 |
AKNAD1
|
AKNA domain containing 1 |
| chr22_+_45714672 | 0.86 |
ENST00000424557.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr19_-_51334769 | 0.85 |
ENST00000596931.1
ENST00000416184.1 ENST00000301421.2 ENST00000598239.1 |
KLK15
|
kallikrein-related peptidase 15 |
| chr5_+_158690089 | 0.85 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr14_-_75536182 | 0.84 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr9_-_35812140 | 0.84 |
ENST00000497810.1
ENST00000396638.2 ENST00000484764.1 |
SPAG8
|
sperm associated antigen 8 |
| chr6_+_24775153 | 0.83 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr16_+_68678892 | 0.81 |
ENST00000429102.2
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr8_-_11996586 | 0.80 |
ENST00000333796.3
|
USP17L2
|
ubiquitin specific peptidase 17-like family member 2 |
| chr15_-_102463298 | 0.80 |
ENST00000326183.3
|
OR4F4
|
olfactory receptor, family 4, subfamily F, member 4 |
| chr1_+_113010056 | 0.79 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr2_+_27371866 | 0.79 |
ENST00000296096.5
|
TCF23
|
transcription factor 23 |
| chr10_-_100027943 | 0.78 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chr21_-_31864275 | 0.77 |
ENST00000334063.4
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chr18_+_32402321 | 0.76 |
ENST00000587723.1
|
DTNA
|
dystrobrevin, alpha |
| chr2_+_46844290 | 0.75 |
ENST00000238892.3
|
CRIPT
|
cysteine-rich PDZ-binding protein |
| chr11_-_110561721 | 0.74 |
ENST00000357139.3
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr19_+_110679 | 0.69 |
ENST00000318050.3
|
OR4F17
|
olfactory receptor, family 4, subfamily F, member 17 |
| chr6_+_46714654 | 0.68 |
ENST00000565422.1
|
ANKRD66
|
ankyrin repeat domain 66 |
| chr2_+_159651821 | 0.68 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr4_+_48807155 | 0.67 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr12_+_80750652 | 0.67 |
ENST00000550182.1
|
OTOGL
|
otogelin-like |
| chr4_-_153332886 | 0.65 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr4_+_189060573 | 0.65 |
ENST00000332517.3
|
TRIML1
|
tripartite motif family-like 1 |
| chrX_-_24045303 | 0.63 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr2_+_78143006 | 0.63 |
ENST00000443419.1
|
AC073628.1
|
AC073628.1 |
| chr9_-_13175823 | 0.62 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr9_-_35812236 | 0.61 |
ENST00000340291.2
|
SPAG8
|
sperm associated antigen 8 |
| chr1_-_149783914 | 0.61 |
ENST00000369167.1
ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF
|
histone cluster 2, H2bf |
| chrX_+_135618258 | 0.61 |
ENST00000440515.1
ENST00000456412.1 |
VGLL1
|
vestigial like 1 (Drosophila) |
| chr8_+_26240666 | 0.57 |
ENST00000523949.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr11_+_56467864 | 0.56 |
ENST00000312153.1
|
OR9G1
|
olfactory receptor, family 9, subfamily G, member 1 |
| chr20_-_35329063 | 0.56 |
ENST00000422536.1
|
NDRG3
|
NDRG family member 3 |
| chr5_+_36608280 | 0.55 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr15_+_81391740 | 0.54 |
ENST00000561216.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr5_-_150138061 | 0.54 |
ENST00000521533.1
ENST00000424236.1 |
DCTN4
|
dynactin 4 (p62) |
| chr3_+_46616017 | 0.53 |
ENST00000542931.1
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr1_+_244227632 | 0.52 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr6_+_13272904 | 0.52 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr2_-_166060552 | 0.51 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr11_-_59383617 | 0.50 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr2_+_113479063 | 0.50 |
ENST00000327581.4
|
NT5DC4
|
5'-nucleotidase domain containing 4 |
| chr1_+_248097071 | 0.50 |
ENST00000318244.3
|
OR2AJ1
|
olfactory receptor, family 2, subfamily AJ, member 1 |
| chr13_+_112240638 | 0.50 |
ENST00000375713.1
|
RP11-65D24.2
|
HCG2045795; Uncharacterized protein |
| chr12_+_78359999 | 0.50 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr6_+_138266498 | 0.49 |
ENST00000434437.1
ENST00000417800.1 |
RP11-240M16.1
|
RP11-240M16.1 |
| chr4_+_22999152 | 0.48 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr3_+_183814852 | 0.48 |
ENST00000415389.2
|
HTR3E
|
5-hydroxytryptamine (serotonin) receptor 3E, ionotropic |
| chr14_-_50999190 | 0.47 |
ENST00000557390.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr8_+_30244580 | 0.47 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr5_-_150138246 | 0.45 |
ENST00000518015.1
|
DCTN4
|
dynactin 4 (p62) |
| chr6_-_31620403 | 0.44 |
ENST00000451898.1
ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr2_-_166060382 | 0.44 |
ENST00000409101.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr20_+_43835638 | 0.44 |
ENST00000372781.3
ENST00000244069.6 |
SEMG1
|
semenogelin I |
| chr12_+_65996599 | 0.43 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr6_-_31620455 | 0.43 |
ENST00000437771.1
ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6
|
BCL2-associated athanogene 6 |
| chr7_-_130598059 | 0.43 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr1_-_111506562 | 0.43 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr5_-_83016632 | 0.42 |
ENST00000515590.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr19_-_53770972 | 0.40 |
ENST00000311170.4
|
VN1R4
|
vomeronasal 1 receptor 4 |
| chr8_+_40018977 | 0.40 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr10_-_4285835 | 0.40 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr20_+_43849941 | 0.39 |
ENST00000372769.3
|
SEMG2
|
semenogelin II |
| chr11_+_62648357 | 0.38 |
ENST00000541372.1
ENST00000539458.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr6_+_46714680 | 0.38 |
ENST00000536046.1
|
ANKRD66
|
ankyrin repeat domain 66 |
| chr6_-_49931818 | 0.37 |
ENST00000322066.3
|
DEFB114
|
defensin, beta 114 |
| chr11_+_6866883 | 0.37 |
ENST00000299454.4
ENST00000379831.2 |
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5 |
| chr12_-_112450915 | 0.36 |
ENST00000437003.2
ENST00000552374.2 ENST00000550831.3 ENST00000354825.3 ENST00000549537.2 ENST00000355445.3 |
TMEM116
|
transmembrane protein 116 |
| chr1_-_108786689 | 0.33 |
ENST00000415641.3
|
NBPF4
|
neuroblastoma breakpoint family, member 4 |
| chr11_-_33795893 | 0.32 |
ENST00000526785.1
ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3
|
F-box protein 3 |
| chr6_+_37897735 | 0.32 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr16_+_76668895 | 0.31 |
ENST00000512279.1
|
RP11-96P7.1
|
RP11-96P7.1 |
| chr2_-_166060571 | 0.31 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr7_-_33140498 | 0.31 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr3_-_126327398 | 0.30 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr4_+_26165074 | 0.28 |
ENST00000512351.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr8_+_94767072 | 0.27 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr3_-_15140629 | 0.27 |
ENST00000507357.1
ENST00000449050.1 ENST00000253699.3 ENST00000435849.3 ENST00000476527.2 |
ZFYVE20
|
zinc finger, FYVE domain containing 20 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.6 | 2.5 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.5 | 2.2 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.5 | 1.6 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.5 | 2.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 2.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 4.1 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.4 | 1.5 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.4 | 9.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.4 | 1.8 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.3 | 1.2 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.3 | 3.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 0.8 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 0.9 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.2 | 0.9 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.2 | 1.6 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.2 | 2.0 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.2 | 2.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.2 | 0.8 | GO:0060901 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.2 | 1.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.2 | 1.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 1.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.8 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.2 | 2.8 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 3.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 1.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.6 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 2.8 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.1 | 0.8 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 1.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.7 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.1 | 1.6 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 1.0 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 2.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 2.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 6.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.6 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 1.1 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.1 | 2.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.9 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 1.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.8 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 1.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 1.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 1.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 1.4 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.9 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 1.9 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.1 | 2.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 1.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.0 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 1.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 2.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.3 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 1.0 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 1.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.5 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 2.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.3 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 1.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 1.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 1.1 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 2.2 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 1.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.7 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 9.9 | GO:0030849 | autosome(GO:0030849) |
| 0.9 | 6.6 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.3 | 1.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 3.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 1.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.2 | 1.6 | GO:0042025 | host cell nucleus(GO:0042025) |
| 0.2 | 1.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 2.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 2.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 1.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 1.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.9 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.6 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 3.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 1.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 4.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 2.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.7 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.7 | 2.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.5 | 2.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.4 | 2.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.4 | 4.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 1.6 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.3 | 6.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 0.9 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.3 | 1.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.3 | 1.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.3 | 1.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.3 | 1.7 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 1.6 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.2 | 2.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 1.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 2.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 2.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 2.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.5 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.9 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 4.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.5 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 1.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.9 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 2.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.8 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 2.3 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 2.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 2.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 5.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.6 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 2.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 4.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 1.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 3.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |