Project
Illumina Body Map 2
Navigation
Downloads
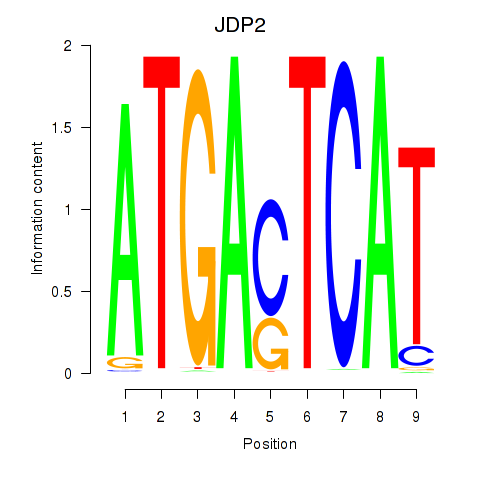
Results for JDP2
Z-value: 1.11
Transcription factors associated with JDP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JDP2
|
ENSG00000140044.8 | Jun dimerization protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JDP2 | hg19_v2_chr14_+_75894714_75894749 | -0.20 | 2.7e-01 | Click! |
Activity profile of JDP2 motif
Sorted Z-values of JDP2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_97361388 | 3.78 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chr20_-_62129163 | 3.71 |
ENST00000298049.7
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr7_+_97361218 | 3.32 |
ENST00000319273.5
|
TAC1
|
tachykinin, precursor 1 |
| chr5_-_41794663 | 3.07 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr5_-_41794313 | 2.68 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr10_-_21463116 | 2.47 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr9_+_34958254 | 2.43 |
ENST00000242315.3
|
KIAA1045
|
KIAA1045 |
| chr2_-_175712479 | 2.42 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr2_-_175711978 | 2.42 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr9_+_34957477 | 2.42 |
ENST00000544237.1
|
KIAA1045
|
KIAA1045 |
| chr2_-_175712270 | 2.37 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr2_-_220173685 | 2.32 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr10_+_88428206 | 2.31 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr16_+_57662138 | 2.31 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr12_+_7023491 | 2.22 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr9_-_23826298 | 2.18 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr14_-_51562037 | 2.08 |
ENST00000338969.5
|
TRIM9
|
tripartite motif containing 9 |
| chr12_+_7023735 | 2.00 |
ENST00000538763.1
ENST00000544774.1 ENST00000545045.2 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr6_+_56820018 | 2.00 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chr1_-_111148241 | 1.86 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr14_-_75083313 | 1.85 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr10_+_88428370 | 1.83 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr6_+_56819773 | 1.77 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr19_-_36019123 | 1.77 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr3_+_155860751 | 1.73 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr7_+_28725585 | 1.67 |
ENST00000396298.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr10_-_75385711 | 1.67 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr8_+_79503458 | 1.67 |
ENST00000518467.1
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr16_+_57662419 | 1.65 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr4_-_176812842 | 1.58 |
ENST00000507540.1
|
GPM6A
|
glycoprotein M6A |
| chr13_-_36429763 | 1.55 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chrX_-_154493791 | 1.55 |
ENST00000369454.3
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr20_-_17539456 | 1.50 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr13_-_67802549 | 1.49 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr8_-_110988070 | 1.49 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr11_-_84028180 | 1.48 |
ENST00000280241.8
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr12_-_131200810 | 1.46 |
ENST00000536002.1
ENST00000544034.1 |
RIMBP2
RP11-662M24.2
|
RIMS binding protein 2 RP11-662M24.2 |
| chr5_+_176237478 | 1.44 |
ENST00000329542.4
|
UNC5A
|
unc-5 homolog A (C. elegans) |
| chr2_-_224467002 | 1.43 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr10_+_134901388 | 1.42 |
ENST00000392607.3
|
GPR123
|
G protein-coupled receptor 123 |
| chr7_-_15014398 | 1.41 |
ENST00000437998.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr12_-_71182695 | 1.38 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr3_-_98241760 | 1.37 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr9_+_44867571 | 1.34 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr19_+_38880695 | 1.34 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr7_-_14942283 | 1.34 |
ENST00000402815.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr1_-_166944652 | 1.31 |
ENST00000528703.1
ENST00000525740.1 ENST00000529387.1 ENST00000469934.2 ENST00000529071.1 ENST00000526687.1 |
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr7_+_147830776 | 1.29 |
ENST00000538075.1
|
CNTNAP2
|
contactin associated protein-like 2 |
| chr11_-_133715394 | 1.29 |
ENST00000299140.3
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chrX_-_20074895 | 1.27 |
ENST00000543767.1
|
MAP7D2
|
MAP7 domain containing 2 |
| chr4_+_26578293 | 1.25 |
ENST00000512840.1
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr9_-_35112376 | 1.23 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr3_+_183894566 | 1.20 |
ENST00000439647.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr18_+_74207477 | 1.19 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chr2_+_27505260 | 1.17 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr6_-_119031228 | 1.15 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr17_-_10701241 | 1.15 |
ENST00000578763.1
|
LINC00675
|
long intergenic non-protein coding RNA 675 |
| chr14_-_51561784 | 1.15 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr20_+_4667094 | 1.14 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr5_+_169758393 | 1.13 |
ENST00000521471.1
ENST00000518357.1 ENST00000436248.3 |
CTB-114C7.3
|
CTB-114C7.3 |
| chr17_-_70417365 | 1.13 |
ENST00000580948.1
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr21_+_22519416 | 1.12 |
ENST00000535285.1
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr3_-_98241713 | 1.12 |
ENST00000502288.1
ENST00000512147.1 ENST00000510541.1 ENST00000503621.1 ENST00000511081.1 |
CLDND1
|
claudin domain containing 1 |
| chr11_-_84028339 | 1.10 |
ENST00000398301.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr16_+_89988259 | 1.10 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr19_-_56056888 | 1.08 |
ENST00000592464.1
ENST00000420723.3 |
SBK3
|
SH3 domain binding kinase family, member 3 |
| chr3_-_98241598 | 1.06 |
ENST00000513287.1
ENST00000514537.1 ENST00000508071.1 ENST00000507944.1 |
CLDND1
|
claudin domain containing 1 |
| chr10_-_116418053 | 1.06 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr16_+_57662527 | 1.05 |
ENST00000563374.1
ENST00000568234.1 ENST00000565770.1 ENST00000564338.1 ENST00000566164.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_-_244006528 | 1.04 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr4_+_120056939 | 1.04 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr6_+_56819895 | 1.00 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr2_+_166326157 | 0.98 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr11_-_57417405 | 0.97 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr6_-_161695042 | 0.97 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr4_+_118349545 | 0.96 |
ENST00000422145.3
ENST00000437514.1 |
AC092661.1
|
AC092661.1 |
| chr2_+_208104351 | 0.96 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr16_+_15596123 | 0.96 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr6_-_44400720 | 0.94 |
ENST00000595057.1
|
AL133262.1
|
AL133262.1 |
| chr4_+_170581213 | 0.93 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr3_-_98241358 | 0.91 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr1_-_166944561 | 0.91 |
ENST00000271417.3
|
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr11_-_82708519 | 0.90 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr10_+_1102303 | 0.88 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr20_+_4666882 | 0.88 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr2_-_145275211 | 0.87 |
ENST00000462355.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr19_-_46285646 | 0.87 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr19_+_49467232 | 0.85 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chr6_-_49755019 | 0.85 |
ENST00000304801.3
|
PGK2
|
phosphoglycerate kinase 2 |
| chr7_-_7603957 | 0.85 |
ENST00000608807.1
|
RP5-1159O4.1
|
RP5-1159O4.1 |
| chr19_-_35992780 | 0.83 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr3_+_183894737 | 0.83 |
ENST00000432591.1
ENST00000431779.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr11_+_26495447 | 0.81 |
ENST00000531568.1
|
ANO3
|
anoctamin 3 |
| chr2_-_145275228 | 0.81 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr10_+_127661942 | 0.81 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr9_-_91793675 | 0.81 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr16_-_14109841 | 0.81 |
ENST00000576797.1
ENST00000575424.1 |
CTD-2135D7.5
|
CTD-2135D7.5 |
| chr8_-_141774467 | 0.80 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr1_+_213224572 | 0.79 |
ENST00000543470.1
ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr16_+_57662596 | 0.78 |
ENST00000567397.1
ENST00000568979.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_+_30077055 | 0.78 |
ENST00000564595.2
ENST00000569798.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr17_+_80317121 | 0.78 |
ENST00000333437.4
|
TEX19
|
testis expressed 19 |
| chr9_+_123906331 | 0.77 |
ENST00000431571.1
|
CNTRL
|
centriolin |
| chr3_+_138153451 | 0.77 |
ENST00000389567.4
ENST00000289135.4 |
ESYT3
|
extended synaptotagmin-like protein 3 |
| chr4_+_71588372 | 0.76 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr3_-_191000172 | 0.73 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr2_+_7073174 | 0.72 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr6_-_161695074 | 0.72 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr9_-_34662651 | 0.71 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr3_+_69134080 | 0.70 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr2_+_87754887 | 0.69 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr1_-_108231101 | 0.69 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr11_+_20620946 | 0.68 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr7_+_102290772 | 0.67 |
ENST00000507450.1
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chr17_-_42994283 | 0.67 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr16_+_9449445 | 0.67 |
ENST00000564305.1
|
RP11-243A14.1
|
RP11-243A14.1 |
| chr3_+_98699880 | 0.66 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr16_+_30077098 | 0.65 |
ENST00000395240.3
ENST00000566846.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr3_-_122512619 | 0.64 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr2_+_87754989 | 0.64 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr7_-_80551671 | 0.63 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr9_-_124991124 | 0.63 |
ENST00000394319.4
ENST00000340587.3 |
LHX6
|
LIM homeobox 6 |
| chr2_-_145275109 | 0.62 |
ENST00000431672.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_154378049 | 0.62 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr17_+_79651007 | 0.62 |
ENST00000572392.1
ENST00000577012.1 |
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr6_+_72596406 | 0.61 |
ENST00000491071.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr4_-_36246060 | 0.61 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr17_+_76311791 | 0.61 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr9_-_123639304 | 0.60 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr12_-_131118370 | 0.60 |
ENST00000541840.1
|
RP11-662M24.2
|
RP11-662M24.2 |
| chr10_+_52152766 | 0.59 |
ENST00000596442.1
|
AC069547.2
|
Uncharacterized protein |
| chr12_-_10324716 | 0.59 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr3_-_45957088 | 0.59 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr8_-_123139423 | 0.58 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr11_-_82708435 | 0.58 |
ENST00000525117.1
ENST00000532548.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr4_+_11627248 | 0.54 |
ENST00000510095.1
|
RP11-281P23.2
|
RP11-281P23.2 |
| chr7_+_127233689 | 0.53 |
ENST00000265825.5
ENST00000420086.2 |
FSCN3
|
fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus) |
| chr3_-_45957534 | 0.53 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_-_246580705 | 0.53 |
ENST00000541742.1
|
SMYD3
|
SET and MYND domain containing 3 |
| chr8_+_70404996 | 0.53 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr7_+_102191679 | 0.52 |
ENST00000507918.1
ENST00000432940.1 |
SPDYE2
|
speedy/RINGO cell cycle regulator family member E2 |
| chr3_+_48507210 | 0.52 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr11_-_57417367 | 0.52 |
ENST00000534810.1
|
YPEL4
|
yippee-like 4 (Drosophila) |
| chr17_-_55162360 | 0.51 |
ENST00000576871.1
ENST00000576313.1 |
RP11-166P13.3
|
RP11-166P13.3 |
| chr15_-_43882140 | 0.51 |
ENST00000429176.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr3_+_69134124 | 0.50 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr1_+_154377669 | 0.50 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr2_+_233562015 | 0.50 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr16_+_30953696 | 0.49 |
ENST00000566320.2
ENST00000565939.1 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr11_+_44069531 | 0.49 |
ENST00000378832.1
|
ACCSL
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional)-like |
| chr17_+_75446629 | 0.49 |
ENST00000588958.2
ENST00000586128.1 |
SEPT9
|
septin 9 |
| chr14_+_86401039 | 0.49 |
ENST00000557195.1
|
CTD-2341M24.1
|
CTD-2341M24.1 |
| chr18_-_74839891 | 0.48 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr5_+_177540444 | 0.47 |
ENST00000274605.5
|
N4BP3
|
NEDD4 binding protein 3 |
| chr6_+_122720681 | 0.47 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chrX_+_1733876 | 0.46 |
ENST00000381241.3
|
ASMT
|
acetylserotonin O-methyltransferase |
| chr6_-_41673552 | 0.46 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr8_-_42623747 | 0.45 |
ENST00000534622.1
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr1_-_95007193 | 0.45 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr12_+_48152774 | 0.45 |
ENST00000549243.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr4_+_175839551 | 0.44 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr12_+_75760714 | 0.44 |
ENST00000547144.1
|
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chr3_+_32023232 | 0.42 |
ENST00000360311.4
|
ZNF860
|
zinc finger protein 860 |
| chr1_-_235116495 | 0.42 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr17_+_66245341 | 0.42 |
ENST00000577985.1
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr22_-_43398442 | 0.42 |
ENST00000422336.1
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr17_-_8263538 | 0.41 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr4_+_175839506 | 0.41 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr12_+_10365082 | 0.41 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr11_-_66104237 | 0.40 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr19_-_51872233 | 0.40 |
ENST00000601435.1
ENST00000291715.1 |
CLDND2
|
claudin domain containing 2 |
| chr17_-_73775839 | 0.39 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr4_+_76481258 | 0.39 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr11_+_28724129 | 0.39 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chrX_+_133371077 | 0.39 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr4_+_22999152 | 0.38 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr11_+_58874701 | 0.38 |
ENST00000529618.1
ENST00000534403.1 ENST00000343597.3 |
FAM111B
|
family with sequence similarity 111, member B |
| chr9_-_123638633 | 0.38 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr8_-_42623924 | 0.38 |
ENST00000276410.2
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr10_+_121410882 | 0.37 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr12_+_10365404 | 0.37 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr2_+_46706725 | 0.37 |
ENST00000434431.1
|
TMEM247
|
transmembrane protein 247 |
| chr17_+_25799008 | 0.37 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr12_-_719573 | 0.37 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr7_-_57207571 | 0.37 |
ENST00000331162.4
|
ZNF479
|
zinc finger protein 479 |
| chr13_+_53602894 | 0.36 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr10_-_4285835 | 0.36 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr6_-_4347271 | 0.35 |
ENST00000437430.2
|
RP3-527G5.1
|
RP3-527G5.1 |
| chr10_-_4285923 | 0.35 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr17_+_79650962 | 0.35 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr21_-_36421626 | 0.35 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr8_-_130253482 | 0.34 |
ENST00000509893.2
|
LINC00977
|
long intergenic non-protein coding RNA 977 |
| chr11_+_122526383 | 0.33 |
ENST00000284273.5
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B |
| chr6_-_31745085 | 0.33 |
ENST00000375686.3
ENST00000447450.1 |
VWA7
|
von Willebrand factor A domain containing 7 |
| chrX_+_41193407 | 0.32 |
ENST00000457138.2
ENST00000441189.2 |
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr17_+_48712138 | 0.32 |
ENST00000515707.1
|
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr21_-_36421535 | 0.32 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr20_-_48782639 | 0.32 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr5_+_145583107 | 0.31 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr11_+_58874658 | 0.31 |
ENST00000411426.1
|
FAM111B
|
family with sequence similarity 111, member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of JDP2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.8 | 3.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.7 | 5.8 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.6 | 2.3 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.4 | 1.3 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.4 | 2.0 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.4 | 1.1 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.3 | 1.9 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.3 | 1.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.3 | 2.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 4.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.2 | 0.6 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 | 1.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 1.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 0.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 1.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 7.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.6 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 1.0 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.8 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 1.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 0.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.8 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 4.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.7 | GO:0060539 | diaphragm development(GO:0060539) negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 5.3 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.2 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 0.5 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 3.8 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.0 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 2.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 1.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.9 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.6 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 2.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 2.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.5 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 1.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 1.0 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.4 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.4 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.8 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 1.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.6 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 1.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 1.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.8 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.2 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.7 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 2.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.3 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.0 | 0.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 3.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 2.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 3.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 1.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 4.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 2.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.0 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 1.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.8 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 3.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 6.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.8 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.4 | 3.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 1.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.3 | 2.0 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.3 | 0.8 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.2 | 1.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 0.7 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 0.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.7 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 6.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 4.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.6 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 3.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 5.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.0 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 4.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 2.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.5 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 1.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 1.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 2.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 2.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 2.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.3 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 2.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 2.9 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 7.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 2.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 5.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 5.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 2.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.0 | 1.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 7.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 4.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 3.7 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |