Project
Illumina Body Map 2
Navigation
Downloads
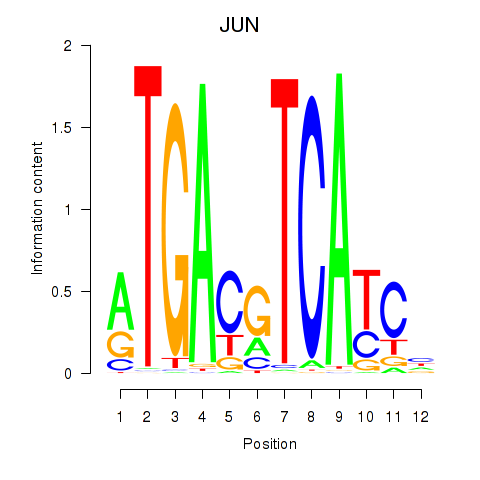
Results for JUN
Z-value: 1.98
Transcription factors associated with JUN
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JUN
|
ENSG00000177606.5 | Jun proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JUN | hg19_v2_chr1_-_59249732_59249785 | 0.30 | 9.8e-02 | Click! |
Activity profile of JUN motif
Sorted Z-values of JUN motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_18891045 | 11.96 |
ENST00000317658.3
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr14_-_55738788 | 9.43 |
ENST00000556183.1
|
RP11-665C16.6
|
RP11-665C16.6 |
| chr12_-_18890940 | 9.15 |
ENST00000543242.1
ENST00000539072.1 ENST00000541966.1 ENST00000266505.7 ENST00000447925.2 ENST00000435379.1 |
PLCZ1
|
phospholipase C, zeta 1 |
| chr2_+_46706725 | 8.92 |
ENST00000434431.1
|
TMEM247
|
transmembrane protein 247 |
| chr14_+_21498360 | 8.74 |
ENST00000321760.6
ENST00000460647.2 ENST00000530140.2 ENST00000472458.1 |
TPPP2
|
tubulin polymerization-promoting protein family member 2 |
| chr9_+_111624577 | 8.48 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr1_+_153175900 | 8.44 |
ENST00000368747.1
|
LELP1
|
late cornified envelope-like proline-rich 1 |
| chr19_+_49109990 | 8.38 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr2_+_232457569 | 8.34 |
ENST00000313965.2
|
C2orf57
|
chromosome 2 open reading frame 57 |
| chr18_+_54814288 | 8.28 |
ENST00000585477.1
|
BOD1L2
|
biorientation of chromosomes in cell division 1-like 2 |
| chr13_+_111972980 | 8.00 |
ENST00000283547.1
|
TEX29
|
testis expressed 29 |
| chr3_-_51909600 | 7.36 |
ENST00000446461.1
|
IQCF5
|
IQ motif containing F5 |
| chr2_+_19911450 | 7.20 |
ENST00000432822.1
|
AC019055.1
|
AC019055.1 |
| chr2_+_171640291 | 7.05 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr20_-_29978383 | 6.91 |
ENST00000339144.3
ENST00000376321.3 |
DEFB119
|
defensin, beta 119 |
| chr18_+_11490077 | 6.78 |
ENST00000586947.1
ENST00000591431.1 |
RP11-712C7.1
|
RP11-712C7.1 |
| chr6_+_97010424 | 6.63 |
ENST00000541107.1
ENST00000450218.1 ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chr12_+_48876275 | 6.53 |
ENST00000314014.2
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr19_-_50979981 | 6.47 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr14_+_21498666 | 6.43 |
ENST00000481535.1
|
TPPP2
|
tubulin polymerization-promoting protein family member 2 |
| chr1_+_110655050 | 6.12 |
ENST00000334179.3
|
UBL4B
|
ubiquitin-like 4B |
| chr19_-_35719609 | 6.11 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187, member B |
| chr9_+_36169380 | 6.05 |
ENST00000335119.2
|
CCIN
|
calicin |
| chr13_+_113030625 | 6.00 |
ENST00000283550.3
|
SPACA7
|
sperm acrosome associated 7 |
| chr5_+_118965244 | 5.87 |
ENST00000515256.1
ENST00000509264.1 |
FAM170A
|
family with sequence similarity 170, member A |
| chr2_-_27362263 | 5.83 |
ENST00000432962.1
ENST00000335524.3 |
C2orf53
|
chromosome 2 open reading frame 53 |
| chr17_-_34270697 | 5.83 |
ENST00000585556.1
|
LYZL6
|
lysozyme-like 6 |
| chr12_-_88423164 | 5.82 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr19_-_4902877 | 5.74 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr19_+_44220162 | 5.69 |
ENST00000244314.5
|
IRGC
|
immunity-related GTPase family, cinema |
| chr20_+_55099542 | 5.50 |
ENST00000371328.3
|
FAM209A
|
family with sequence similarity 209, member A |
| chr1_+_40713573 | 5.49 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr9_-_27297126 | 5.20 |
ENST00000380031.1
ENST00000537675.1 ENST00000380032.3 |
EQTN
|
equatorin, sperm acrosome associated |
| chr17_-_34270668 | 5.17 |
ENST00000293274.4
|
LYZL6
|
lysozyme-like 6 |
| chr19_-_49250054 | 5.15 |
ENST00000602105.1
ENST00000332955.2 |
IZUMO1
|
izumo sperm-egg fusion 1 |
| chr19_-_8809139 | 5.13 |
ENST00000324436.3
|
ACTL9
|
actin-like 9 |
| chr19_+_44220247 | 5.02 |
ENST00000596627.1
|
IRGC
|
immunity-related GTPase family, cinema |
| chr1_-_76398077 | 5.02 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr3_-_51937331 | 4.99 |
ENST00000310914.5
|
IQCF1
|
IQ motif containing F1 |
| chr11_-_5531215 | 4.97 |
ENST00000311659.4
|
UBQLN3
|
ubiquilin 3 |
| chrX_+_102965835 | 4.94 |
ENST00000319560.6
|
TMEM31
|
transmembrane protein 31 |
| chr12_-_13256571 | 4.88 |
ENST00000545401.1
ENST00000432710.2 ENST00000351606.6 |
GSG1
|
germ cell associated 1 |
| chr3_+_44840679 | 4.81 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr20_+_42295745 | 4.74 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr19_+_36024310 | 4.73 |
ENST00000222286.4
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr6_+_42123141 | 4.72 |
ENST00000418175.1
ENST00000541991.1 ENST00000053469.4 ENST00000394237.1 ENST00000372963.1 |
GUCA1A
RP1-139D8.6
|
guanylate cyclase activator 1A (retina) RP1-139D8.6 |
| chr10_+_22634384 | 4.60 |
ENST00000376624.3
ENST00000376603.2 ENST00000376601.1 ENST00000538630.1 ENST00000456231.2 ENST00000313311.6 ENST00000435326.1 |
SPAG6
|
sperm associated antigen 6 |
| chrX_-_139047669 | 4.58 |
ENST00000370540.1
|
CXorf66
|
chromosome X open reading frame 66 |
| chr3_+_51863433 | 4.51 |
ENST00000444293.1
|
IQCF3
|
IQ motif containing F3 |
| chr9_-_113100088 | 4.48 |
ENST00000374510.4
ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa) |
| chr22_+_51176624 | 4.47 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chrX_+_83116142 | 4.44 |
ENST00000329312.4
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr7_-_37956409 | 4.38 |
ENST00000436072.2
|
SFRP4
|
secreted frizzled-related protein 4 |
| chr3_-_195938256 | 4.35 |
ENST00000296326.3
|
ZDHHC19
|
zinc finger, DHHC-type containing 19 |
| chr16_-_11367452 | 4.32 |
ENST00000327157.2
|
PRM3
|
protamine 3 |
| chr7_-_121944491 | 4.31 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr14_-_44976474 | 4.29 |
ENST00000340446.4
|
FSCB
|
fibrous sheath CABYR binding protein |
| chrY_-_20935572 | 4.28 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr12_-_13256593 | 4.25 |
ENST00000542415.1
ENST00000324458.8 |
GSG1
|
germ cell associated 1 |
| chr9_+_105757590 | 4.25 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chrY_+_20708557 | 4.24 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr1_+_34632484 | 4.23 |
ENST00000373374.3
|
C1orf94
|
chromosome 1 open reading frame 94 |
| chr9_-_34397800 | 4.22 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr2_+_3705785 | 4.20 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr1_-_120439112 | 4.19 |
ENST00000369400.1
|
ADAM30
|
ADAM metallopeptidase domain 30 |
| chr2_-_61389168 | 4.18 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr20_+_55108302 | 4.11 |
ENST00000371325.1
|
FAM209B
|
family with sequence similarity 209, member B |
| chr13_+_46276441 | 4.10 |
ENST00000310521.1
ENST00000533564.1 |
SPERT
|
spermatid associated |
| chr20_+_43343886 | 4.09 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr19_-_55874611 | 4.08 |
ENST00000424985.3
|
FAM71E2
|
family with sequence similarity 71, member E2 |
| chr4_-_147043058 | 4.08 |
ENST00000512063.1
ENST00000507726.1 |
RP11-6L6.3
|
long intergenic non-protein coding RNA 1095 |
| chr12_+_54378923 | 4.06 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr7_+_151653464 | 4.06 |
ENST00000431418.2
ENST00000392800.2 |
GALNTL5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr7_+_142636603 | 4.05 |
ENST00000409607.3
|
C7orf34
|
chromosome 7 open reading frame 34 |
| chr11_-_5531159 | 3.98 |
ENST00000445998.1
|
UBQLN3
|
ubiquilin 3 |
| chr20_-_29978286 | 3.97 |
ENST00000376315.2
|
DEFB119
|
defensin, beta 119 |
| chr20_+_34203794 | 3.97 |
ENST00000374273.3
|
SPAG4
|
sperm associated antigen 4 |
| chr17_+_31318886 | 3.95 |
ENST00000269053.3
ENST00000394638.1 |
SPACA3
|
sperm acrosome associated 3 |
| chr1_+_1115056 | 3.93 |
ENST00000379288.3
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10 |
| chr3_+_51851612 | 3.89 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr20_-_44420507 | 3.83 |
ENST00000243938.4
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr16_-_3545424 | 3.81 |
ENST00000437192.3
ENST00000399645.3 |
C16orf90
|
chromosome 16 open reading frame 90 |
| chrY_+_3447082 | 3.73 |
ENST00000321217.4
ENST00000559055.2 |
TGIF2LY
|
TGFB-induced factor homeobox 2-like, Y-linked |
| chr1_+_64669294 | 3.72 |
ENST00000371077.5
|
UBE2U
|
ubiquitin-conjugating enzyme E2U (putative) |
| chrX_-_154689596 | 3.70 |
ENST00000369444.2
|
H2AFB3
|
H2A histone family, member B3 |
| chr7_+_73275483 | 3.68 |
ENST00000320531.2
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28 |
| chr7_+_142636440 | 3.60 |
ENST00000458732.1
|
C7orf34
|
chromosome 7 open reading frame 34 |
| chr12_-_119199461 | 3.60 |
ENST00000536572.1
ENST00000542577.1 |
RP11-3L23.2
|
RP11-3L23.2 |
| chr19_-_50666431 | 3.55 |
ENST00000293405.3
|
IZUMO2
|
IZUMO family member 2 |
| chr15_+_76016293 | 3.54 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chrX_+_151867214 | 3.52 |
ENST00000329342.5
ENST00000412733.1 ENST00000457643.1 |
MAGEA6
|
melanoma antigen family A, 6 |
| chr1_-_162346657 | 3.51 |
ENST00000367935.5
|
C1orf111
|
chromosome 1 open reading frame 111 |
| chr15_-_80634104 | 3.46 |
ENST00000561432.1
ENST00000558913.1 ENST00000558578.1 |
LINC00927
|
long intergenic non-protein coding RNA 927 |
| chrX_+_89176881 | 3.42 |
ENST00000283891.5
ENST00000561129.2 |
TGIF2LX
|
TGFB-induced factor homeobox 2-like, X-linked |
| chr11_-_65793948 | 3.39 |
ENST00000312106.5
|
CATSPER1
|
cation channel, sperm associated 1 |
| chr17_-_8661860 | 3.37 |
ENST00000328794.6
|
SPDYE4
|
speedy/RINGO cell cycle regulator family member E4 |
| chr17_+_33448593 | 3.34 |
ENST00000158009.5
|
FNDC8
|
fibronectin type III domain containing 8 |
| chr16_-_52640834 | 3.33 |
ENST00000510238.3
|
CASC16
|
cancer susceptibility candidate 16 (non-protein coding) |
| chr12_-_52604607 | 3.30 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr13_-_60737898 | 3.30 |
ENST00000377908.2
ENST00000400319.1 ENST00000400320.1 ENST00000267215.4 |
DIAPH3
|
diaphanous-related formin 3 |
| chr8_-_18711866 | 3.27 |
ENST00000519851.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr11_-_68780824 | 3.27 |
ENST00000441623.1
ENST00000309099.6 |
MRGPRF
|
MAS-related GPR, member F |
| chr22_-_44258360 | 3.26 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr1_-_95783809 | 3.24 |
ENST00000423410.1
|
RP4-586O15.1
|
RP4-586O15.1 |
| chr22_-_17073700 | 3.21 |
ENST00000359963.3
|
CCT8L2
|
chaperonin containing TCP1, subunit 8 (theta)-like 2 |
| chr22_+_50528308 | 3.21 |
ENST00000545383.1
ENST00000262794.5 |
MOV10L1
|
Mov10l1, Moloney leukemia virus 10-like 1, homolog (mouse) |
| chr2_-_61389240 | 3.14 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr12_-_49999417 | 3.13 |
ENST00000257894.2
|
FAM186B
|
family with sequence similarity 186, member B |
| chr11_-_28129656 | 3.10 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr14_+_77292715 | 3.09 |
ENST00000393774.3
ENST00000555189.1 ENST00000450042.2 |
C14orf166B
|
chromosome 14 open reading frame 166B |
| chr9_-_19033237 | 3.05 |
ENST00000380530.1
ENST00000542071.1 |
FAM154A
|
family with sequence similarity 154, member A |
| chr10_-_131762105 | 2.96 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr5_-_147286065 | 2.95 |
ENST00000318315.4
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr9_-_19033185 | 2.94 |
ENST00000380534.4
|
FAM154A
|
family with sequence similarity 154, member A |
| chr20_+_32254286 | 2.92 |
ENST00000330271.4
|
ACTL10
|
actin-like 10 |
| chr2_+_74011316 | 2.91 |
ENST00000409561.1
|
C2orf78
|
chromosome 2 open reading frame 78 |
| chr22_+_50528459 | 2.87 |
ENST00000395858.3
ENST00000395843.1 |
MOV10L1
|
Mov10l1, Moloney leukemia virus 10-like 1, homolog (mouse) |
| chr20_+_23471727 | 2.86 |
ENST00000449810.1
ENST00000246012.1 |
CST8
|
cystatin 8 (cystatin-related epididymal specific) |
| chr17_+_39994032 | 2.86 |
ENST00000293303.4
ENST00000438813.1 |
KLHL10
|
kelch-like family member 10 |
| chr17_-_63822563 | 2.83 |
ENST00000317442.8
|
CEP112
|
centrosomal protein 112kDa |
| chr4_+_186347388 | 2.82 |
ENST00000511138.1
ENST00000511581.1 |
C4orf47
|
chromosome 4 open reading frame 47 |
| chr6_-_112575912 | 2.82 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr17_+_41005283 | 2.81 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chr4_+_128802016 | 2.81 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr1_-_222014008 | 2.78 |
ENST00000431729.1
|
RP11-191N8.2
|
RP11-191N8.2 |
| chr1_-_26701003 | 2.78 |
ENST00000455900.1
|
ZNF683
|
zinc finger protein 683 |
| chr12_-_49999389 | 2.77 |
ENST00000551047.1
ENST00000544141.1 |
FAM186B
|
family with sequence similarity 186, member B |
| chr15_+_74528630 | 2.76 |
ENST00000398814.3
|
CCDC33
|
coiled-coil domain containing 33 |
| chr20_+_17680587 | 2.74 |
ENST00000427254.1
ENST00000377805.3 |
BANF2
|
barrier to autointegration factor 2 |
| chr12_-_4754339 | 2.71 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr22_+_22988816 | 2.68 |
ENST00000480559.1
ENST00000448514.1 |
GGTLC2
|
gamma-glutamyltransferase light chain 2 |
| chrX_+_92929192 | 2.67 |
ENST00000332647.4
|
FAM133A
|
family with sequence similarity 133, member A |
| chr1_+_85527987 | 2.64 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr19_-_50666404 | 2.63 |
ENST00000600293.1
|
IZUMO2
|
IZUMO family member 2 |
| chr4_+_94125100 | 2.62 |
ENST00000512631.1
|
GRID2
|
glutamate receptor, ionotropic, delta 2 |
| chr1_+_96457545 | 2.61 |
ENST00000413825.2
|
RP11-147C23.1
|
Uncharacterized protein |
| chr6_-_136571400 | 2.58 |
ENST00000418509.2
ENST00000420702.1 ENST00000451457.2 |
MTFR2
|
mitochondrial fission regulator 2 |
| chr20_-_31124186 | 2.58 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr16_+_30034655 | 2.58 |
ENST00000300575.2
|
C16orf92
|
chromosome 16 open reading frame 92 |
| chr19_+_7011509 | 2.56 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr1_-_32264356 | 2.55 |
ENST00000452755.2
|
SPOCD1
|
SPOC domain containing 1 |
| chr6_+_144185573 | 2.54 |
ENST00000237275.6
ENST00000539295.1 |
ZC2HC1B
|
zinc finger, C2HC-type containing 1B |
| chr20_+_17680599 | 2.54 |
ENST00000246090.5
|
BANF2
|
barrier to autointegration factor 2 |
| chr14_+_60386853 | 2.49 |
ENST00000570145.1
|
LRRC9
|
leucine rich repeat containing 9 |
| chr17_+_61562201 | 2.47 |
ENST00000290863.6
ENST00000413513.3 ENST00000421982.2 |
ACE
|
angiotensin I converting enzyme |
| chr5_-_139944196 | 2.46 |
ENST00000357560.4
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr5_+_39520499 | 2.45 |
ENST00000604954.1
|
CTD-2078B5.2
|
CTD-2078B5.2 |
| chr5_-_99870932 | 2.44 |
ENST00000504833.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr13_+_113030658 | 2.42 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr1_-_32264250 | 2.42 |
ENST00000528579.1
|
SPOCD1
|
SPOC domain containing 1 |
| chr1_-_178840157 | 2.40 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr3_+_126113734 | 2.40 |
ENST00000352312.1
ENST00000393425.1 |
CCDC37
|
coiled-coil domain containing 37 |
| chr14_+_93389425 | 2.35 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr7_+_142031986 | 2.35 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr12_-_4754356 | 2.35 |
ENST00000540967.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr17_-_56621665 | 2.35 |
ENST00000321691.3
|
C17orf47
|
chromosome 17 open reading frame 47 |
| chr12_-_4754318 | 2.32 |
ENST00000536414.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr17_+_12569306 | 2.32 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr4_-_104119528 | 2.32 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr9_-_34665983 | 2.32 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr11_+_117070037 | 2.31 |
ENST00000392951.4
ENST00000525531.1 ENST00000278968.6 |
TAGLN
|
transgelin |
| chrX_-_152160740 | 2.31 |
ENST00000361887.5
ENST00000439251.1 ENST00000452693.1 |
PNMA5
|
paraneoplastic Ma antigen family member 5 |
| chr11_+_6226782 | 2.30 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr6_-_112575758 | 2.29 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr3_-_21792838 | 2.28 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chrX_+_47863734 | 2.26 |
ENST00000304355.5
|
SPACA5
|
sperm acrosome associated 5 |
| chrX_-_151922340 | 2.26 |
ENST00000370284.1
ENST00000543232.1 ENST00000393876.1 ENST00000393872.3 |
MAGEA2
|
melanoma antigen family A, 2 |
| chr17_-_76899275 | 2.25 |
ENST00000322630.2
ENST00000586713.1 |
DDC8
|
Protein DDC8 homolog |
| chrX_+_154610428 | 2.21 |
ENST00000354514.4
|
H2AFB2
|
H2A histone family, member B2 |
| chr18_+_11490028 | 2.19 |
ENST00000591318.1
|
RP11-712C7.1
|
RP11-712C7.1 |
| chr18_+_76829258 | 2.18 |
ENST00000588600.1
|
ATP9B
|
ATPase, class II, type 9B |
| chrX_+_154113317 | 2.18 |
ENST00000354461.2
|
H2AFB1
|
H2A histone family, member B1 |
| chr15_-_80634126 | 2.16 |
ENST00000558244.1
|
LINC00927
|
long intergenic non-protein coding RNA 927 |
| chr1_+_55107449 | 2.16 |
ENST00000421030.2
ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7
|
maestro heat-like repeat family member 7 |
| chr6_+_28048753 | 2.16 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr9_+_130478345 | 2.15 |
ENST00000373289.3
ENST00000393748.4 |
TTC16
|
tetratricopeptide repeat domain 16 |
| chr2_+_197577841 | 2.12 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr4_-_186456652 | 2.12 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr2_+_38177620 | 2.12 |
ENST00000402091.3
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr4_-_186456766 | 2.10 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr20_+_43343517 | 2.08 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr17_+_61562178 | 2.08 |
ENST00000490216.2
|
ACE
|
angiotensin I converting enzyme |
| chr18_+_11490061 | 2.07 |
ENST00000589566.1
|
RP11-712C7.1
|
RP11-712C7.1 |
| chr22_+_19118321 | 2.07 |
ENST00000399635.2
|
TSSK2
|
testis-specific serine kinase 2 |
| chr10_-_50342053 | 2.06 |
ENST00000311787.5
|
FAM170B
|
family with sequence similarity 170, member B |
| chr1_+_173837488 | 2.04 |
ENST00000427304.1
ENST00000432989.1 ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr12_-_52887034 | 2.03 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr12_-_100660833 | 2.02 |
ENST00000551642.1
ENST00000416321.1 ENST00000550587.1 ENST00000549249.1 |
DEPDC4
|
DEP domain containing 4 |
| chr20_+_43343476 | 2.02 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chrX_+_151883090 | 2.01 |
ENST00000370293.2
ENST00000423993.1 ENST00000447530.1 ENST00000458057.1 ENST00000331220.2 ENST00000422085.1 ENST00000453150.1 ENST00000409560.1 |
MAGEA2B
|
melanoma antigen family A, 2B |
| chr14_+_23025534 | 2.00 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr8_-_19093121 | 1.98 |
ENST00000518417.1
|
RP11-1080G15.1
|
RP11-1080G15.1 |
| chr3_+_158288999 | 1.96 |
ENST00000482628.1
ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr16_+_11343475 | 1.96 |
ENST00000572173.1
|
RMI2
|
RecQ mediated genome instability 2 |
| chrX_-_154060946 | 1.95 |
ENST00000369529.1
|
SMIM9
|
small integral membrane protein 9 |
| chrX_-_101771645 | 1.92 |
ENST00000289373.4
|
TMSB15A
|
thymosin beta 15a |
| chr8_-_10336885 | 1.91 |
ENST00000520494.1
|
RP11-981G7.3
|
RP11-981G7.3 |
| chr5_-_145483932 | 1.90 |
ENST00000311450.4
|
PLAC8L1
|
PLAC8-like 1 |
| chr11_-_2160180 | 1.90 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr17_+_7835419 | 1.89 |
ENST00000576538.1
ENST00000380262.3 ENST00000563694.1 ENST00000380255.3 ENST00000570782.1 |
CNTROB
|
centrobin, centrosomal BRCA2 interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of JUN
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 34.0 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.7 | 8.6 | GO:0007343 | egg activation(GO:0007343) |
| 1.5 | 4.4 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.1 | 4.2 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 1.0 | 5.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 1.0 | 2.0 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 1.0 | 5.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.9 | 2.8 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.9 | 4.5 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.9 | 3.6 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.8 | 2.4 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.8 | 2.4 | GO:2000705 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.7 | 4.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.7 | 4.2 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.7 | 6.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.6 | 1.9 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.6 | 3.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.6 | 2.9 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.6 | 4.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.5 | 2.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.5 | 3.4 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.5 | 1.4 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.4 | 0.9 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.4 | 3.8 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.4 | 1.7 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.4 | 6.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.4 | 2.8 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.4 | 2.8 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.4 | 1.5 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.4 | 1.5 | GO:0032771 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.4 | 2.6 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.4 | 1.1 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.4 | 1.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.4 | 1.1 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.4 | 3.9 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.3 | 2.4 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.3 | 5.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.3 | 1.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 0.9 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.3 | 0.9 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.3 | 1.6 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.3 | 0.9 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.3 | 5.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 1.1 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.3 | 0.8 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.3 | 12.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.3 | 1.9 | GO:0060283 | growth plate cartilage chondrocyte proliferation(GO:0003419) negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.3 | 0.5 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.2 | 1.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.2 | 6.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 4.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 2.2 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 1.9 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.2 | 1.9 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 2.7 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 0.8 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.2 | 0.8 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 0.8 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.2 | 3.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.2 | 1.5 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.2 | 4.1 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.2 | 1.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 0.9 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.2 | 0.7 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.5 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.2 | 1.9 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 7.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 1.7 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 0.7 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 1.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 4.7 | GO:0051197 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.2 | 0.5 | GO:0042214 | terpene metabolic process(GO:0042214) phytoalexin metabolic process(GO:0052314) |
| 0.2 | 0.5 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.2 | 0.6 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 1.1 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 4.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.6 | GO:0045844 | positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) |
| 0.1 | 1.4 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 2.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 1.6 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 2.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 9.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 3.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 3.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 4.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.3 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 4.9 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 2.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 1.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.4 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 12.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 5.8 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 0.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.3 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.2 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.6 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 1.6 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.1 | 1.7 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.4 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 0.4 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 1.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 9.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.3 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 0.4 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 1.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.3 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 0.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.7 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.9 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 | 3.2 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 0.3 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.2 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 1.5 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 6.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 2.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.3 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 1.1 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.1 | 1.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.6 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 2.4 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.8 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 1.1 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.9 | GO:0035150 | regulation of tube size(GO:0035150) regulation of blood vessel size(GO:0050880) |
| 0.0 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 2.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.9 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 1.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 1.8 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 1.0 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 0.5 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 3.5 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.0 | 0.3 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 1.4 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 4.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 4.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 4.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.4 | GO:1903522 | regulation of blood circulation(GO:1903522) |
| 0.0 | 0.4 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 0.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 1.6 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 17.5 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.4 | GO:0010611 | regulation of cardiac muscle hypertrophy(GO:0010611) regulation of muscle hypertrophy(GO:0014743) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 1.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 2.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 1.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.8 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.6 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.3 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 2.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.6 | GO:0090175 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 | 1.0 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 1.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.5 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.9 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) |
| 0.0 | 7.0 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.3 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0060754 | regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.5 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.8 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.2 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 1.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.3 | GO:0009798 | axis specification(GO:0009798) |
| 0.0 | 0.3 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.3 | GO:0050953 | visual perception(GO:0007601) sensory perception of light stimulus(GO:0050953) |
| 0.0 | 0.2 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 2.2 | 4.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.5 | 7.3 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 1.3 | 5.0 | GO:0031523 | Myb complex(GO:0031523) |
| 1.2 | 4.8 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.8 | 11.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.6 | 12.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.6 | 7.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.6 | 6.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.5 | 8.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.5 | 1.4 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.4 | 5.6 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.4 | 0.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.4 | 2.8 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 3.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 3.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.3 | 1.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 0.9 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 4.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 4.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 10.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.3 | 0.9 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.3 | 0.8 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 1.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 21.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 0.9 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 1.9 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 1.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 1.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 1.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 4.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 0.5 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 3.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.9 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 11.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 4.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 2.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.6 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 1.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 2.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 4.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 0.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 2.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 5.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 3.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 1.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 13.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 4.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 2.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 10.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 2.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 1.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.7 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 34.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.2 | 4.7 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.1 | 4.5 | GO:0004040 | amidase activity(GO:0004040) |
| 1.1 | 3.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.7 | 2.8 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.7 | 4.7 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.6 | 5.2 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.4 | 3.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 1.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.4 | 1.8 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.4 | 3.9 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.3 | 3.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.3 | 1.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.3 | 0.9 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.3 | 1.9 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.3 | 4.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 0.9 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.3 | 2.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.3 | 2.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 4.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 1.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 1.9 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.2 | 0.8 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.2 | 1.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 5.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 1.7 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.2 | 4.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) S-adenosyl-L-methionine binding(GO:1904047) |
| 0.2 | 4.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 6.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 2.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 4.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 5.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 3.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 1.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 5.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.3 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 4.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.3 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.1 | 0.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 7.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 1.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 3.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.4 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 3.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 2.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.5 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.1 | 1.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 2.6 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 3.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 13.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 7.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 5.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 18.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 4.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 4.0 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 2.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 2.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 1.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 4.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 2.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 2.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 4.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 14.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.3 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 9.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0010857 | calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 1.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.0 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.0 | GO:0016730 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 2.2 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 10.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 21.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 4.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 2.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 2.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 3.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 4.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 4.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 7.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 3.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 4.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 7.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.8 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.9 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.9 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 3.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |