Project
Illumina Body Map 2
Navigation
Downloads
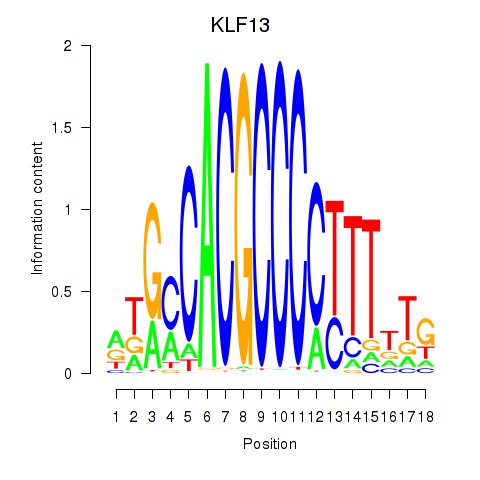
Results for KLF13
Z-value: 0.91
Transcription factors associated with KLF13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF13
|
ENSG00000169926.5 | Kruppel like factor 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF13 | hg19_v2_chr15_+_31619013_31619095 | 0.37 | 3.6e-02 | Click! |
Activity profile of KLF13 motif
Sorted Z-values of KLF13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_27068704 | 5.37 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr19_-_7936344 | 2.16 |
ENST00000599142.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr15_+_30375158 | 1.88 |
ENST00000341650.6
ENST00000567927.1 |
GOLGA8J
|
golgin A8 family, member J |
| chr15_+_41851211 | 1.82 |
ENST00000263798.3
|
TYRO3
|
TYRO3 protein tyrosine kinase |
| chr6_+_30594619 | 1.67 |
ENST00000318999.7
ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1
|
alpha tubulin acetyltransferase 1 |
| chr19_-_19729725 | 1.67 |
ENST00000251203.9
|
PBX4
|
pre-B-cell leukemia homeobox 4 |
| chr7_-_99774945 | 1.66 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr15_+_40453204 | 1.65 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr22_+_27068766 | 1.59 |
ENST00000435162.1
ENST00000437071.1 ENST00000440816.1 ENST00000421253.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr19_+_49867181 | 1.47 |
ENST00000597546.1
|
DKKL1
|
dickkopf-like 1 |
| chrX_-_140271249 | 1.44 |
ENST00000370526.2
|
LDOC1
|
leucine zipper, down-regulated in cancer 1 |
| chr14_-_67981916 | 1.39 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr15_+_30896329 | 1.35 |
ENST00000566740.1
|
GOLGA8H
|
golgin A8 family, member H |
| chr1_+_33352036 | 1.34 |
ENST00000373467.3
|
HPCA
|
hippocalcin |
| chr12_+_49961990 | 1.30 |
ENST00000551063.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr6_-_11779403 | 1.28 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr19_+_49866851 | 1.28 |
ENST00000221498.2
ENST00000596402.1 |
DKKL1
|
dickkopf-like 1 |
| chrX_+_15756382 | 1.26 |
ENST00000318636.3
|
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chrX_-_72097698 | 1.20 |
ENST00000373530.1
|
DMRTC1
|
DMRT-like family C1 |
| chrX_+_72062802 | 1.18 |
ENST00000373533.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr19_-_55672037 | 1.14 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr7_+_5632436 | 1.14 |
ENST00000340250.6
ENST00000382361.3 |
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| chr2_-_241500168 | 1.13 |
ENST00000443318.1
ENST00000411765.1 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr19_+_39616410 | 1.13 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr19_+_19649057 | 1.12 |
ENST00000586018.1
ENST00000291495.5 |
CILP2
|
cartilage intermediate layer protein 2 |
| chr19_+_49866331 | 1.10 |
ENST00000597873.1
|
DKKL1
|
dickkopf-like 1 |
| chr21_+_15588499 | 1.09 |
ENST00000400577.3
|
RBM11
|
RNA binding motif protein 11 |
| chr6_-_31697563 | 1.07 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr1_-_27709793 | 1.06 |
ENST00000374027.3
ENST00000374025.3 |
CD164L2
|
CD164 sialomucin-like 2 |
| chr1_+_21766641 | 1.05 |
ENST00000342104.5
|
NBPF3
|
neuroblastoma breakpoint family, member 3 |
| chr19_-_22715280 | 1.05 |
ENST00000593802.1
|
ZNF98
|
zinc finger protein 98 |
| chr19_+_7701985 | 1.05 |
ENST00000595950.1
ENST00000441779.2 ENST00000221283.5 ENST00000414284.2 |
STXBP2
|
syntaxin binding protein 2 |
| chrX_+_72062617 | 1.02 |
ENST00000440247.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr12_-_6580094 | 1.00 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr11_-_94964354 | 0.99 |
ENST00000536441.1
|
SESN3
|
sestrin 3 |
| chr15_-_28957469 | 0.98 |
ENST00000563027.1
ENST00000340249.3 |
GOLGA8M
|
golgin A8 family, member M |
| chr2_-_239140011 | 0.98 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr19_+_1071203 | 0.97 |
ENST00000543365.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr4_+_55524085 | 0.97 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr13_+_24734880 | 0.96 |
ENST00000382095.4
|
SPATA13
|
spermatogenesis associated 13 |
| chr12_+_58013693 | 0.95 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr15_+_23255242 | 0.94 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr6_-_33285505 | 0.93 |
ENST00000431845.2
|
ZBTB22
|
zinc finger and BTB domain containing 22 |
| chr11_-_94964210 | 0.93 |
ENST00000416495.2
ENST00000393234.1 |
SESN3
|
sestrin 3 |
| chr15_+_90792760 | 0.92 |
ENST00000339615.5
ENST00000438251.1 |
TTLL13
|
tubulin tyrosine ligase-like family, member 13 |
| chr6_-_11779014 | 0.92 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr12_-_6579808 | 0.91 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr12_-_6579833 | 0.90 |
ENST00000396308.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr2_+_114737146 | 0.90 |
ENST00000412690.1
|
AC010982.1
|
AC010982.1 |
| chr1_-_27709816 | 0.89 |
ENST00000374030.1
|
CD164L2
|
CD164 sialomucin-like 2 |
| chr19_-_14629224 | 0.89 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr1_-_244615425 | 0.89 |
ENST00000366535.3
|
ADSS
|
adenylosuccinate synthase |
| chr1_+_874649 | 0.88 |
ENST00000455979.1
|
SAMD11
|
sterile alpha motif domain containing 11 |
| chr5_+_170288856 | 0.88 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr2_-_239140276 | 0.87 |
ENST00000334973.4
|
AC016757.3
|
Protein LOC151174 |
| chr7_-_155089251 | 0.87 |
ENST00000609974.1
|
AC144652.1
|
AC144652.1 |
| chr1_-_42921915 | 0.86 |
ENST00000372565.3
ENST00000433602.2 |
ZMYND12
|
zinc finger, MYND-type containing 12 |
| chr12_-_6982442 | 0.86 |
ENST00000523102.1
ENST00000524270.1 ENST00000519357.1 |
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr1_+_21766588 | 0.86 |
ENST00000454000.2
ENST00000318220.6 ENST00000318249.5 |
NBPF3
|
neuroblastoma breakpoint family, member 3 |
| chr7_+_99775366 | 0.85 |
ENST00000394018.2
ENST00000416412.1 |
STAG3
|
stromal antigen 3 |
| chr6_+_41040678 | 0.84 |
ENST00000341376.6
ENST00000353205.5 |
NFYA
|
nuclear transcription factor Y, alpha |
| chr7_-_99756293 | 0.84 |
ENST00000316937.3
ENST00000456769.1 |
C7orf43
|
chromosome 7 open reading frame 43 |
| chr15_-_30686052 | 0.83 |
ENST00000562729.1
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr6_-_11779174 | 0.83 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr7_+_99775520 | 0.83 |
ENST00000317296.5
ENST00000422690.1 ENST00000439782.1 |
STAG3
|
stromal antigen 3 |
| chr17_-_19281203 | 0.82 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr20_+_37590942 | 0.81 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr20_-_23402028 | 0.81 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chrX_+_134555863 | 0.80 |
ENST00000417443.2
|
LINC00086
|
long intergenic non-protein coding RNA 86 |
| chr16_-_28936493 | 0.79 |
ENST00000544477.1
ENST00000357573.6 |
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chrX_-_103401649 | 0.78 |
ENST00000357421.4
|
SLC25A53
|
solute carrier family 25, member 53 |
| chr19_+_5455421 | 0.77 |
ENST00000222033.4
|
ZNRF4
|
zinc and ring finger 4 |
| chr14_+_68286478 | 0.77 |
ENST00000487861.1
|
RAD51B
|
RAD51 paralog B |
| chrX_+_48911090 | 0.76 |
ENST00000597275.1
|
CCDC120
|
coiled-coil domain containing 120 |
| chr6_+_31126291 | 0.76 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr15_+_72947079 | 0.75 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family, member B |
| chr1_+_15573757 | 0.74 |
ENST00000358897.4
ENST00000417793.1 ENST00000375999.3 ENST00000433640.2 |
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr2_-_85645545 | 0.73 |
ENST00000409275.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr4_+_1795012 | 0.72 |
ENST00000481110.2
ENST00000340107.4 ENST00000440486.2 ENST00000412135.2 |
FGFR3
|
fibroblast growth factor receptor 3 |
| chr19_-_12941469 | 0.71 |
ENST00000586969.1
ENST00000589681.1 ENST00000585384.1 ENST00000589808.1 |
RTBDN
|
retbindin |
| chr15_+_23599993 | 0.69 |
ENST00000562295.1
|
GOLGA8S
|
golgin A8 family, member S |
| chr4_-_13546632 | 0.69 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr15_+_75575176 | 0.68 |
ENST00000434739.3
|
GOLGA6D
|
golgin A6 family, member D |
| chr11_+_65383227 | 0.68 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr17_+_27055798 | 0.68 |
ENST00000268766.6
|
NEK8
|
NIMA-related kinase 8 |
| chr1_-_223316577 | 0.68 |
ENST00000407096.2
|
TLR5
|
toll-like receptor 5 |
| chr16_-_28936007 | 0.67 |
ENST00000568703.1
ENST00000567483.1 |
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr15_-_30685752 | 0.66 |
ENST00000299847.2
ENST00000397827.3 |
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr1_-_16939976 | 0.66 |
ENST00000430580.2
|
NBPF1
|
neuroblastoma breakpoint family, member 1 |
| chr14_-_67981870 | 0.65 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr9_+_91926103 | 0.64 |
ENST00000314355.6
|
CKS2
|
CDC28 protein kinase regulatory subunit 2 |
| chr19_-_46145696 | 0.63 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr15_-_74374891 | 0.63 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr3_-_138048631 | 0.62 |
ENST00000484930.1
ENST00000475751.1 |
NME9
|
NME/NM23 family member 9 |
| chr21_+_48055527 | 0.62 |
ENST00000397638.2
ENST00000458387.2 ENST00000451211.2 ENST00000291705.6 ENST00000397637.1 ENST00000334494.4 ENST00000397628.1 ENST00000440086.1 |
PRMT2
|
protein arginine methyltransferase 2 |
| chr19_-_10764509 | 0.61 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr6_-_31697255 | 0.61 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_+_58938903 | 0.60 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr5_+_111496554 | 0.58 |
ENST00000442823.2
|
EPB41L4A-AS1
|
EPB41L4A antisense RNA 1 |
| chr19_-_47164386 | 0.57 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr4_-_137020391 | 0.57 |
ENST00000569694.1
|
RP11-775H9.3
|
RP11-775H9.3 |
| chr19_+_36249057 | 0.56 |
ENST00000301165.5
ENST00000536950.1 ENST00000537459.1 ENST00000421853.2 |
C19orf55
|
chromosome 19 open reading frame 55 |
| chr11_+_65479702 | 0.56 |
ENST00000530446.1
ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr11_+_123986069 | 0.55 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr19_+_7985198 | 0.55 |
ENST00000221573.6
ENST00000595637.1 |
SNAPC2
|
small nuclear RNA activating complex, polypeptide 2, 45kDa |
| chr1_-_146082633 | 0.55 |
ENST00000605317.1
ENST00000604938.1 ENST00000339388.5 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr1_+_172502336 | 0.55 |
ENST00000263688.3
|
SUCO
|
SUN domain containing ossification factor |
| chr18_+_20513278 | 0.54 |
ENST00000327155.5
|
RBBP8
|
retinoblastoma binding protein 8 |
| chr20_+_3776371 | 0.54 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr12_+_6982725 | 0.53 |
ENST00000433346.1
|
LRRC23
|
leucine rich repeat containing 23 |
| chr11_+_7041677 | 0.53 |
ENST00000299481.4
|
NLRP14
|
NLR family, pyrin domain containing 14 |
| chr1_+_39547082 | 0.53 |
ENST00000602421.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr7_+_100450328 | 0.53 |
ENST00000540482.1
ENST00000418037.1 ENST00000428758.1 ENST00000275729.3 ENST00000415287.1 ENST00000354161.3 ENST00000416675.1 |
SLC12A9
|
solute carrier family 12, member 9 |
| chr11_-_33722286 | 0.52 |
ENST00000451594.2
ENST00000379011.4 |
C11orf91
|
chromosome 11 open reading frame 91 |
| chr17_-_7165662 | 0.52 |
ENST00000571881.2
ENST00000360325.7 |
CLDN7
|
claudin 7 |
| chr19_+_36249044 | 0.51 |
ENST00000444637.2
ENST00000396908.4 ENST00000544099.1 |
C19orf55
|
chromosome 19 open reading frame 55 |
| chr19_+_8509842 | 0.51 |
ENST00000325495.4
ENST00000600092.1 ENST00000594907.1 ENST00000596984.1 ENST00000601645.1 |
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M |
| chr19_+_10765003 | 0.51 |
ENST00000407004.3
ENST00000589998.1 ENST00000589600.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr1_+_45805728 | 0.50 |
ENST00000539779.1
|
TOE1
|
target of EGR1, member 1 (nuclear) |
| chr4_+_9174899 | 0.49 |
ENST00000432515.1
|
FAM90A26
|
family with sequence similarity 90, member A26 |
| chr2_-_160143059 | 0.48 |
ENST00000392796.3
|
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr14_+_68286516 | 0.48 |
ENST00000471583.1
ENST00000487270.1 ENST00000488612.1 |
RAD51B
|
RAD51 paralog B |
| chr2_-_160143084 | 0.47 |
ENST00000409990.3
|
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chrX_+_26156460 | 0.46 |
ENST00000325250.1
|
MAGEB18
|
melanoma antigen family B, 18 |
| chr2_-_160143242 | 0.45 |
ENST00000359774.4
|
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr2_+_179316163 | 0.45 |
ENST00000409117.3
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr7_+_86273218 | 0.45 |
ENST00000361669.2
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chr11_-_89653576 | 0.45 |
ENST00000420869.1
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr4_-_76439483 | 0.44 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr7_+_12727250 | 0.43 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr6_-_169364429 | 0.43 |
ENST00000444586.1
|
RP3-495K2.3
|
RP3-495K2.3 |
| chr1_+_39546988 | 0.42 |
ENST00000484793.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr19_+_46236509 | 0.41 |
ENST00000457052.2
|
AC074212.3
|
Uncharacterized protein |
| chr1_-_45805752 | 0.40 |
ENST00000354383.6
ENST00000355498.2 ENST00000372100.5 ENST00000531105.1 |
MUTYH
|
mutY homolog |
| chr15_-_32695396 | 0.40 |
ENST00000512626.2
ENST00000435655.2 |
GOLGA8K
AC139426.2
|
golgin A8 family, member K Uncharacterized protein; cDNA FLJ52611 |
| chr15_-_20747114 | 0.40 |
ENST00000427390.2
|
GOLGA6L6
|
golgin A6 family-like 6 |
| chr9_+_132597722 | 0.40 |
ENST00000372429.3
ENST00000315480.4 ENST00000358355.1 |
USP20
|
ubiquitin specific peptidase 20 |
| chr15_+_22736246 | 0.40 |
ENST00000316397.3
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chr7_-_111846435 | 0.40 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr2_-_160143158 | 0.39 |
ENST00000409124.1
ENST00000358147.4 |
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr1_-_45805607 | 0.39 |
ENST00000372104.1
ENST00000448481.1 ENST00000483127.1 ENST00000528013.2 ENST00000456914.2 |
MUTYH
|
mutY homolog |
| chr15_-_65117807 | 0.39 |
ENST00000559239.1
ENST00000268043.4 ENST00000333425.6 |
PIF1
|
PIF1 5'-to-3' DNA helicase |
| chr6_+_52535878 | 0.39 |
ENST00000211314.4
|
TMEM14A
|
transmembrane protein 14A |
| chr18_-_51750948 | 0.38 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr16_-_89883015 | 0.37 |
ENST00000563673.1
ENST00000389301.3 ENST00000568369.1 ENST00000534992.1 ENST00000389302.3 ENST00000543736.1 |
FANCA
|
Fanconi anemia, complementation group A |
| chr15_+_22736484 | 0.37 |
ENST00000560659.2
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chr2_+_114737472 | 0.37 |
ENST00000420161.1
|
AC110769.3
|
AC110769.3 |
| chr1_-_19283163 | 0.36 |
ENST00000455833.2
|
IFFO2
|
intermediate filament family orphan 2 |
| chrX_+_54835493 | 0.36 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr1_+_172502244 | 0.36 |
ENST00000610051.1
|
SUCO
|
SUN domain containing ossification factor |
| chr19_+_41284121 | 0.35 |
ENST00000594800.1
ENST00000357052.2 ENST00000602173.1 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr2_-_241500447 | 0.35 |
ENST00000536462.1
ENST00000405002.1 ENST00000441168.1 ENST00000403283.1 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr19_+_10764937 | 0.35 |
ENST00000449870.1
ENST00000318511.3 ENST00000420083.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr15_+_32885657 | 0.35 |
ENST00000448387.2
ENST00000569659.1 |
GOLGA8N
|
golgin A8 family, member N |
| chr17_+_42264322 | 0.34 |
ENST00000446571.3
ENST00000357984.3 ENST00000538716.2 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr19_+_45844018 | 0.34 |
ENST00000585434.1
|
KLC3
|
kinesin light chain 3 |
| chr7_+_155089486 | 0.33 |
ENST00000340368.4
ENST00000344756.4 ENST00000425172.1 ENST00000342407.5 |
INSIG1
|
insulin induced gene 1 |
| chr12_+_58120044 | 0.33 |
ENST00000542466.2
|
AGAP2-AS1
|
AGAP2 antisense RNA 1 |
| chr7_+_100210133 | 0.33 |
ENST00000393950.2
ENST00000424091.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr1_+_65775204 | 0.33 |
ENST00000371069.4
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr19_-_6767516 | 0.33 |
ENST00000245908.6
|
SH2D3A
|
SH2 domain containing 3A |
| chr19_+_45843994 | 0.32 |
ENST00000391946.2
|
KLC3
|
kinesin light chain 3 |
| chr7_+_12726623 | 0.32 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr7_+_100209725 | 0.31 |
ENST00000223054.4
|
MOSPD3
|
motile sperm domain containing 3 |
| chr7_+_100209979 | 0.31 |
ENST00000493970.1
ENST00000379527.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr7_+_111846741 | 0.31 |
ENST00000421043.1
ENST00000425229.1 ENST00000450657.1 |
ZNF277
|
zinc finger protein 277 |
| chr2_+_71295766 | 0.31 |
ENST00000533981.1
|
NAGK
|
N-acetylglucosamine kinase |
| chr1_+_45805342 | 0.31 |
ENST00000372090.5
|
TOE1
|
target of EGR1, member 1 (nuclear) |
| chr6_-_105307417 | 0.30 |
ENST00000524020.1
|
HACE1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr4_+_699610 | 0.30 |
ENST00000521023.2
|
PCGF3
|
polycomb group ring finger 3 |
| chr10_+_104678102 | 0.30 |
ENST00000433628.2
|
CNNM2
|
cyclin M2 |
| chr11_-_76341399 | 0.29 |
ENST00000532413.1
|
RP11-672A2.5
|
RP11-672A2.5 |
| chr14_+_24867992 | 0.29 |
ENST00000382554.3
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr17_-_40333150 | 0.29 |
ENST00000264661.3
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chrX_-_13062752 | 0.29 |
ENST00000380625.3
ENST00000542843.1 |
FAM9C
|
family with sequence similarity 9, member C |
| chr12_-_10095997 | 0.29 |
ENST00000412084.1
|
AC091814.2
|
AC091814.2 |
| chr5_-_32313082 | 0.29 |
ENST00000382142.3
|
MTMR12
|
myotubularin related protein 12 |
| chr15_+_41056218 | 0.28 |
ENST00000260447.4
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr7_+_2281843 | 0.27 |
ENST00000356714.1
ENST00000397049.1 |
NUDT1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr6_-_31697977 | 0.27 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr12_+_105724613 | 0.27 |
ENST00000549934.2
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr11_+_64685026 | 0.26 |
ENST00000526559.1
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr19_-_6767431 | 0.26 |
ENST00000437152.3
ENST00000597687.1 |
SH2D3A
|
SH2 domain containing 3A |
| chr6_-_41040268 | 0.26 |
ENST00000373154.2
ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr19_-_36236292 | 0.25 |
ENST00000378975.3
ENST00000412391.2 ENST00000292879.5 |
U2AF1L4
|
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr15_+_84904525 | 0.25 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr22_+_21771656 | 0.25 |
ENST00000407464.2
|
HIC2
|
hypermethylated in cancer 2 |
| chr13_+_27131887 | 0.25 |
ENST00000335327.5
|
WASF3
|
WAS protein family, member 3 |
| chr15_-_28778117 | 0.24 |
ENST00000525590.2
ENST00000329523.6 |
GOLGA8G
|
golgin A8 family, member G |
| chr16_-_30905263 | 0.24 |
ENST00000572628.1
|
BCL7C
|
B-cell CLL/lymphoma 7C |
| chr1_-_45806053 | 0.24 |
ENST00000412971.1
ENST00000372098.3 ENST00000372110.3 ENST00000529984.1 ENST00000528332.2 ENST00000372115.3 ENST00000450313.1 |
MUTYH
|
mutY homolog |
| chr14_-_23299009 | 0.24 |
ENST00000488800.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr19_-_19843900 | 0.23 |
ENST00000344099.3
|
ZNF14
|
zinc finger protein 14 |
| chr19_-_9420249 | 0.23 |
ENST00000591998.1
|
ZNF699
|
zinc finger protein 699 |
| chr19_-_12941215 | 0.23 |
ENST00000458671.2
|
RTBDN
|
retbindin |
| chr17_-_13505219 | 0.23 |
ENST00000284110.1
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr4_+_4291924 | 0.23 |
ENST00000355834.3
ENST00000337872.4 ENST00000538529.1 ENST00000502918.1 |
ZBTB49
|
zinc finger and BTB domain containing 49 |
| chr6_+_106534192 | 0.22 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr13_+_27131798 | 0.22 |
ENST00000361042.4
|
WASF3
|
WAS protein family, member 3 |
| chr15_-_26874230 | 0.22 |
ENST00000400188.3
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 0.8 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.4 | 1.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.3 | 1.0 | GO:0038162 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.2 | 1.3 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.2 | 0.9 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 1.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 1.2 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.9 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 1.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.5 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 2.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.5 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 1.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 2.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 1.7 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.2 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.4 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.9 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.4 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.3 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 0.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.7 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 1.4 | GO:1900120 | regulation of receptor binding(GO:1900120) |
| 0.0 | 0.4 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.7 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 1.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 3.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.4 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 1.1 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 1.0 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.2 | GO:0006023 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 1.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 1.0 | GO:0007507 | heart development(GO:0007507) |
| 0.0 | 1.7 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 1.0 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.8 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.5 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 1.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.7 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.4 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.3 | GO:0000732 | strand displacement(GO:0000732) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 0.7 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 0.6 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 1.7 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.2 | 1.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.6 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.7 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.9 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 2.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.3 | 0.9 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.3 | 2.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 1.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.1 | 1.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 2.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.4 | GO:0033677 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 0.5 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 1.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.3 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.5 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 1.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 2.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.8 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 1.5 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 1.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |