Project
Illumina Body Map 2
Navigation
Downloads
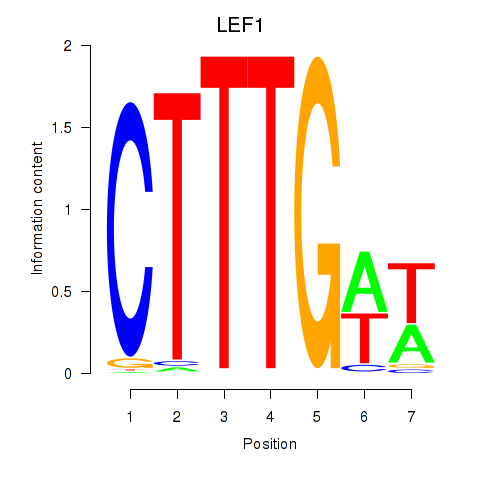
Results for LEF1
Z-value: 1.92
Transcription factors associated with LEF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LEF1
|
ENSG00000138795.5 | lymphoid enhancer binding factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LEF1 | hg19_v2_chr4_-_109087906_109087953 | -0.46 | 7.8e-03 | Click! |
Activity profile of LEF1 motif
Sorted Z-values of LEF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_124553437 | 7.25 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr5_+_140480083 | 7.06 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr10_+_123970670 | 6.63 |
ENST00000496913.2
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr11_-_111794446 | 6.09 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr7_-_27219849 | 5.96 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr1_+_78383813 | 5.56 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr3_+_173302222 | 5.04 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr10_+_54074033 | 4.92 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr3_-_149375783 | 4.68 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr21_+_40823753 | 4.67 |
ENST00000333634.4
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr12_-_16759711 | 4.57 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_+_159141397 | 4.52 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr11_-_41481135 | 4.50 |
ENST00000528697.1
ENST00000530763.1 |
LRRC4C
|
leucine rich repeat containing 4C |
| chr9_-_97356075 | 4.50 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr21_+_40824003 | 4.49 |
ENST00000452550.1
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr6_-_127840453 | 4.33 |
ENST00000556132.1
|
SOGA3
|
SOGA family member 3 |
| chr8_-_124665190 | 4.13 |
ENST00000325995.7
|
KLHL38
|
kelch-like family member 38 |
| chr12_-_16760021 | 4.09 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr4_-_186877502 | 4.09 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_224467093 | 4.06 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr1_+_170633047 | 4.06 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr7_-_27224795 | 4.05 |
ENST00000006015.3
|
HOXA11
|
homeobox A11 |
| chr5_-_146833485 | 3.98 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr15_+_63335899 | 3.94 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr15_+_96869165 | 3.94 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr7_-_27224842 | 3.94 |
ENST00000517402.1
|
HOXA11
|
homeobox A11 |
| chr15_-_94614049 | 3.89 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr9_+_133971909 | 3.88 |
ENST00000247291.3
ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L
|
allograft inflammatory factor 1-like |
| chr2_-_183387064 | 3.81 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr13_-_36429763 | 3.73 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr17_-_63557309 | 3.72 |
ENST00000580513.1
|
AXIN2
|
axin 2 |
| chr15_+_43885252 | 3.71 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr15_+_43985725 | 3.69 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr5_-_146833803 | 3.68 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr7_+_70597109 | 3.65 |
ENST00000333538.5
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17 |
| chr12_-_16759440 | 3.59 |
ENST00000537304.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr7_-_27213893 | 3.59 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr4_+_95972822 | 3.57 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chrX_-_32173579 | 3.57 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chrX_-_15619076 | 3.52 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chrX_+_38420783 | 3.51 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr10_+_123923205 | 3.50 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr7_+_20370746 | 3.47 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr20_+_36149602 | 3.43 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr3_+_2553281 | 3.42 |
ENST00000434053.1
|
CNTN4
|
contactin 4 |
| chr2_+_176994408 | 3.40 |
ENST00000429017.1
ENST00000313173.4 ENST00000544999.1 |
HOXD8
|
homeobox D8 |
| chr3_-_99569821 | 3.39 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr1_-_234667504 | 3.39 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr4_+_20255123 | 3.39 |
ENST00000504154.1
ENST00000273739.5 |
SLIT2
|
slit homolog 2 (Drosophila) |
| chr4_+_150999418 | 3.36 |
ENST00000296550.7
|
DCLK2
|
doublecortin-like kinase 2 |
| chr9_+_133971863 | 3.36 |
ENST00000372309.3
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr7_+_20370300 | 3.34 |
ENST00000537992.1
|
ITGB8
|
integrin, beta 8 |
| chr15_+_43985084 | 3.30 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr12_-_16761007 | 3.29 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr4_-_88449771 | 3.27 |
ENST00000535835.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr2_+_172950227 | 3.26 |
ENST00000341900.6
|
DLX1
|
distal-less homeobox 1 |
| chr3_-_134093395 | 3.25 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr4_-_88450595 | 3.25 |
ENST00000434434.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr5_-_153857819 | 3.22 |
ENST00000231121.2
|
HAND1
|
heart and neural crest derivatives expressed 1 |
| chr7_+_140774032 | 3.20 |
ENST00000565468.1
|
TMEM178B
|
transmembrane protein 178B |
| chr11_+_112832090 | 3.19 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chrX_-_138287168 | 3.16 |
ENST00000436198.1
|
FGF13
|
fibroblast growth factor 13 |
| chr3_+_29323043 | 3.12 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr13_-_110959478 | 3.11 |
ENST00000543140.1
ENST00000375820.4 |
COL4A1
|
collagen, type IV, alpha 1 |
| chr11_-_47207390 | 3.10 |
ENST00000539589.1
ENST00000528462.1 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr1_+_100111479 | 3.09 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chr6_-_105584560 | 3.08 |
ENST00000336775.5
|
BVES
|
blood vessel epicardial substance |
| chr2_-_224467002 | 3.07 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr2_-_175629164 | 3.07 |
ENST00000409323.1
ENST00000261007.5 ENST00000348749.5 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr14_-_23624511 | 3.05 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr8_+_80523321 | 3.04 |
ENST00000518111.1
|
STMN2
|
stathmin-like 2 |
| chr1_+_100111580 | 3.00 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr18_+_32073253 | 2.98 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr12_-_6233828 | 2.97 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr3_+_132757215 | 2.96 |
ENST00000321871.6
ENST00000393130.3 ENST00000514894.1 ENST00000512662.1 |
TMEM108
|
transmembrane protein 108 |
| chr8_-_21988558 | 2.96 |
ENST00000312841.8
|
HR
|
hair growth associated |
| chr7_+_30185406 | 2.95 |
ENST00000324489.5
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr5_+_131593364 | 2.92 |
ENST00000253754.3
ENST00000379018.3 |
PDLIM4
|
PDZ and LIM domain 4 |
| chr10_+_73724123 | 2.90 |
ENST00000373115.4
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3 |
| chr2_-_183387430 | 2.87 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr11_-_8285405 | 2.86 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr7_-_80548493 | 2.82 |
ENST00000536800.1
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr12_+_53491220 | 2.81 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr5_-_44388899 | 2.81 |
ENST00000264664.4
|
FGF10
|
fibroblast growth factor 10 |
| chr7_+_73868439 | 2.78 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr18_-_24445729 | 2.77 |
ENST00000383168.4
|
AQP4
|
aquaporin 4 |
| chr2_-_183387283 | 2.76 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr14_+_69726864 | 2.76 |
ENST00000448469.3
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr1_+_6845384 | 2.75 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr12_-_16762971 | 2.74 |
ENST00000540590.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_+_110009215 | 2.74 |
ENST00000369872.3
|
SYPL2
|
synaptophysin-like 2 |
| chrX_-_128788914 | 2.74 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr8_-_22089845 | 2.73 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr14_-_61124977 | 2.71 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr10_-_87551311 | 2.69 |
ENST00000536331.1
|
GRID1
|
glutamate receptor, ionotropic, delta 1 |
| chrX_-_43832711 | 2.68 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr2_-_86564776 | 2.68 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr2_-_217560248 | 2.66 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr5_-_160973649 | 2.65 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr3_-_187388173 | 2.64 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr20_-_42815733 | 2.62 |
ENST00000342272.3
|
JPH2
|
junctophilin 2 |
| chr1_+_203096831 | 2.61 |
ENST00000337894.4
|
ADORA1
|
adenosine A1 receptor |
| chr6_-_112575912 | 2.60 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr2_+_220309379 | 2.59 |
ENST00000451076.1
|
SPEG
|
SPEG complex locus |
| chr15_-_93632421 | 2.59 |
ENST00000329082.7
|
RGMA
|
repulsive guidance molecule family member a |
| chr2_-_175629135 | 2.59 |
ENST00000409542.1
ENST00000409219.1 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr7_+_73245193 | 2.56 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr12_+_7167980 | 2.56 |
ENST00000360817.5
ENST00000402681.3 |
C1S
|
complement component 1, s subcomponent |
| chr2_-_72374948 | 2.54 |
ENST00000546307.1
ENST00000474509.1 |
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr19_-_18717627 | 2.54 |
ENST00000392386.3
|
CRLF1
|
cytokine receptor-like factor 1 |
| chrX_-_80457385 | 2.54 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr8_-_22089533 | 2.53 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr14_+_69726968 | 2.51 |
ENST00000553669.1
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr3_-_58563094 | 2.51 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr3_-_134093275 | 2.51 |
ENST00000513145.1
ENST00000422605.2 |
AMOTL2
|
angiomotin like 2 |
| chr11_+_112832202 | 2.50 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr11_-_118047376 | 2.47 |
ENST00000278947.5
|
SCN2B
|
sodium channel, voltage-gated, type II, beta subunit |
| chr14_-_21270995 | 2.45 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr6_+_138483058 | 2.43 |
ENST00000251691.4
|
KIAA1244
|
KIAA1244 |
| chr2_-_152382500 | 2.43 |
ENST00000434685.1
|
NEB
|
nebulin |
| chr20_+_10199566 | 2.41 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr5_+_140797296 | 2.38 |
ENST00000398594.2
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr2_-_50574856 | 2.37 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr8_-_17579726 | 2.36 |
ENST00000381861.3
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr16_+_6069586 | 2.36 |
ENST00000547372.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_+_198669365 | 2.35 |
ENST00000428675.1
|
PLCL1
|
phospholipase C-like 1 |
| chr10_+_24497704 | 2.34 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr7_-_27239703 | 2.34 |
ENST00000222753.4
|
HOXA13
|
homeobox A13 |
| chr15_+_80696666 | 2.34 |
ENST00000303329.4
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr21_+_35552978 | 2.33 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr17_-_63557759 | 2.33 |
ENST00000307078.5
|
AXIN2
|
axin 2 |
| chr2_+_14772810 | 2.32 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chr1_+_215256467 | 2.32 |
ENST00000391894.2
ENST00000444842.2 |
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr6_-_127840048 | 2.28 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr6_+_69942915 | 2.22 |
ENST00000604969.1
ENST00000603207.1 |
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr18_-_53089538 | 2.22 |
ENST00000566777.1
|
TCF4
|
transcription factor 4 |
| chr2_-_86564740 | 2.21 |
ENST00000540790.1
ENST00000428491.1 |
REEP1
|
receptor accessory protein 1 |
| chr2_-_72375167 | 2.21 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr4_+_114214125 | 2.20 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr15_-_41120896 | 2.19 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr8_+_85095769 | 2.18 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr20_+_10199468 | 2.18 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr3_-_66024213 | 2.17 |
ENST00000483466.1
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr10_+_24498060 | 2.17 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr12_-_22063787 | 2.16 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chrX_+_135229559 | 2.15 |
ENST00000394155.2
|
FHL1
|
four and a half LIM domains 1 |
| chr9_+_124088860 | 2.14 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr9_-_91793675 | 2.14 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr10_-_116164239 | 2.12 |
ENST00000419268.1
ENST00000304129.4 ENST00000545353.1 |
AFAP1L2
|
actin filament associated protein 1-like 2 |
| chr5_+_140864649 | 2.12 |
ENST00000306593.1
|
PCDHGC4
|
protocadherin gamma subfamily C, 4 |
| chr8_-_119964434 | 2.12 |
ENST00000297350.4
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b |
| chr7_-_80548667 | 2.12 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr10_-_61900762 | 2.11 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr18_-_24445664 | 2.09 |
ENST00000578776.1
|
AQP4
|
aquaporin 4 |
| chr1_+_6845578 | 2.09 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr12_-_16761117 | 2.09 |
ENST00000538051.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_+_101962128 | 2.09 |
ENST00000550514.1
|
MYBPC1
|
myosin binding protein C, slow type |
| chr11_+_113930955 | 2.08 |
ENST00000535700.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr9_-_23825956 | 2.08 |
ENST00000397312.2
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr18_-_31802056 | 2.07 |
ENST00000538587.1
|
NOL4
|
nucleolar protein 4 |
| chr11_-_119234876 | 2.06 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chrX_+_135229600 | 2.06 |
ENST00000370690.3
|
FHL1
|
four and a half LIM domains 1 |
| chr3_+_259218 | 2.05 |
ENST00000449294.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr4_-_5891918 | 2.05 |
ENST00000512574.1
|
CRMP1
|
collapsin response mediator protein 1 |
| chr4_-_20985632 | 2.05 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr16_+_6069072 | 2.04 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr4_+_15341442 | 2.03 |
ENST00000397700.2
ENST00000295297.4 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chrX_+_46937745 | 2.03 |
ENST00000397180.1
ENST00000457380.1 ENST00000352078.4 |
RGN
|
regucalcin |
| chr11_+_101983176 | 2.03 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr2_+_18059906 | 2.01 |
ENST00000304101.4
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr11_+_112832133 | 2.00 |
ENST00000524665.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr14_+_85996471 | 2.00 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr7_+_30185496 | 1.99 |
ENST00000455738.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr20_+_57875457 | 1.99 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chr21_+_19617140 | 1.97 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr3_+_29322437 | 1.97 |
ENST00000434693.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr18_-_31802282 | 1.97 |
ENST00000535475.1
|
NOL4
|
nucleolar protein 4 |
| chr9_+_101705893 | 1.97 |
ENST00000375001.3
|
COL15A1
|
collagen, type XV, alpha 1 |
| chr5_+_175288631 | 1.96 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr1_+_170632250 | 1.96 |
ENST00000367760.3
|
PRRX1
|
paired related homeobox 1 |
| chr19_+_13135439 | 1.96 |
ENST00000586873.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr19_+_30719410 | 1.96 |
ENST00000585628.1
ENST00000591488.1 |
ZNF536
|
zinc finger protein 536 |
| chr1_+_82266053 | 1.93 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr14_+_24837226 | 1.92 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr4_-_5894777 | 1.92 |
ENST00000324989.7
|
CRMP1
|
collapsin response mediator protein 1 |
| chr2_-_161056802 | 1.92 |
ENST00000283249.2
ENST00000409872.1 |
ITGB6
|
integrin, beta 6 |
| chr7_-_27219632 | 1.91 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr17_+_27920486 | 1.91 |
ENST00000394859.3
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr2_+_155555201 | 1.91 |
ENST00000544049.1
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr16_-_4987065 | 1.90 |
ENST00000590782.2
ENST00000345988.2 |
PPL
|
periplakin |
| chr10_-_116164450 | 1.89 |
ENST00000369271.3
|
AFAP1L2
|
actin filament associated protein 1-like 2 |
| chr1_+_215179188 | 1.88 |
ENST00000391895.2
|
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr14_+_85996507 | 1.88 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr12_-_130529501 | 1.87 |
ENST00000561864.1
ENST00000567788.1 |
RP11-474D1.4
RP11-474D1.3
|
RP11-474D1.4 RP11-474D1.3 |
| chr11_-_125351481 | 1.86 |
ENST00000577924.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr3_+_189349162 | 1.83 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr11_-_104769141 | 1.83 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr6_-_127840021 | 1.82 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr20_+_30598231 | 1.82 |
ENST00000300415.8
ENST00000262659.8 |
CCM2L
|
cerebral cavernous malformation 2-like |
| chr11_-_47206965 | 1.82 |
ENST00000525725.1
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of LEF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 1.9 | 5.6 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 1.6 | 4.9 | GO:0100012 | regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 1.6 | 4.8 | GO:2001035 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 1.5 | 20.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.4 | 5.5 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.3 | 6.3 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 1.3 | 5.0 | GO:0097115 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 1.1 | 4.6 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 1.1 | 4.5 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 1.1 | 3.3 | GO:0021893 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 1.1 | 2.1 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 1.1 | 3.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 1.0 | 3.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 1.0 | 3.1 | GO:0060931 | sinoatrial node development(GO:0003163) sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.9 | 2.8 | GO:0071338 | proximal/distal axis specification(GO:0009946) bronchiole development(GO:0060435) submandibular salivary gland formation(GO:0060661) secretion by lung epithelial cell involved in lung growth(GO:0061033) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.9 | 6.4 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.9 | 2.7 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.9 | 4.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.9 | 2.7 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.9 | 1.8 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 0.9 | 2.6 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.8 | 4.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.8 | 4.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.8 | 3.2 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.8 | 1.6 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.8 | 3.1 | GO:0060901 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.8 | 3.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.8 | 3.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.7 | 2.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.7 | 3.5 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.7 | 2.7 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.7 | 2.0 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.7 | 2.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.7 | 3.3 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.6 | 3.9 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.6 | 4.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.6 | 5.7 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.6 | 1.8 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.6 | 5.9 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.6 | 11.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.5 | 3.2 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.5 | 1.5 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.5 | 2.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.5 | 21.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.5 | 3.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.5 | 0.9 | GO:0060197 | cloacal septation(GO:0060197) |
| 0.5 | 1.8 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.4 | 4.9 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.4 | 2.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 0.4 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.4 | 2.1 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.4 | 2.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 5.3 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.4 | 7.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.4 | 1.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.4 | 1.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.4 | 5.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.4 | 1.9 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.4 | 7.5 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.4 | 1.8 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.4 | 8.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 2.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.4 | 2.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 1.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 1.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 1.4 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 6.5 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 1.0 | GO:1903800 | regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.3 | 2.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 12.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.3 | 3.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 0.6 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.3 | 3.7 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.3 | 1.2 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.3 | 7.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 2.3 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.3 | 6.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.3 | 5.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 7.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.3 | 2.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 3.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 1.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 2.6 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.3 | 2.9 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 3.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.3 | 1.0 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.3 | 1.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 6.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 1.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 1.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 1.7 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 1.5 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 1.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 1.0 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.2 | 1.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 11.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 0.8 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.2 | 0.6 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.2 | 3.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 2.3 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.2 | 2.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 1.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 1.0 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.2 | 1.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.2 | 1.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 0.5 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.2 | 2.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 1.9 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.2 | 0.6 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.2 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.2 | 0.8 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.2 | 1.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 0.8 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 1.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 6.7 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 8.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 5.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.6 | GO:0070293 | renal sodium ion transport(GO:0003096) renal absorption(GO:0070293) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.7 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.1 | 2.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 2.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 1.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.5 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 4.9 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.5 | GO:0060462 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.1 | 1.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.6 | GO:0050893 | sensory processing(GO:0050893) |
| 0.1 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.7 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 3.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.3 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 2.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 1.8 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 1.0 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 6.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.6 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 5.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.3 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 1.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.7 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 1.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.6 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 1.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 3.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.8 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 0.3 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.1 | 3.2 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 0.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 2.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 3.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 1.4 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 1.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 1.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 1.3 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.1 | 15.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.6 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.8 | GO:0051828 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.1 | 3.4 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 0.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 4.9 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.4 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 2.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.2 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.1 | 4.0 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.5 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 1.9 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 0.9 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.1 | 0.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.3 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 4.2 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 0.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 5.0 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.9 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 2.1 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.1 | 1.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 3.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 8.4 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 0.4 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 0.3 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 1.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 2.8 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 3.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 8.0 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 1.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.7 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 3.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.9 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 2.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 1.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 8.2 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.9 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.7 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 1.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 2.2 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 2.7 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 2.4 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.9 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.7 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.8 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 1.3 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.8 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 1.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 1.7 | GO:0060047 | heart contraction(GO:0060047) |
| 0.0 | 1.0 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.8 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 2.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 1.0 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 2.5 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.5 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 1.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.8 | GO:0097502 | mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 2.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.4 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.4 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.8 | GO:0032874 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.0 | 0.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.5 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.0 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.0 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 1.6 | 3.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.7 | 3.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.6 | 4.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.5 | 2.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.5 | 5.1 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.5 | 5.5 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.4 | 4.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.4 | 6.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 6.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 3.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.3 | 2.0 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.3 | 8.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 1.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 2.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 6.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 6.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 6.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 1.8 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 5.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 2.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.6 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.2 | 1.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 1.2 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 3.8 | GO:0032059 | bleb(GO:0032059) |
| 0.2 | 1.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 10.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 6.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 6.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 3.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 39.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 2.0 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 2.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 5.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 9.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.4 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 1.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.6 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 1.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 3.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 2.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 9.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 7.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 2.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 3.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 3.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 9.7 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 0.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 8.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 3.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 17.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 28.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 11.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 4.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 4.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 9.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 6.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 3.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 6.3 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 4.2 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.4 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.1 | 6.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.1 | 4.5 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 1.1 | 3.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.9 | 6.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.8 | 4.1 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.8 | 3.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.7 | 3.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.7 | 7.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 4.8 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.6 | 7.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.5 | 2.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.5 | 6.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.5 | 2.6 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.4 | 3.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 1.6 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.4 | 1.2 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.4 | 5.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 6.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.4 | 3.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 1.1 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.4 | 2.9 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 2.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.3 | 3.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 2.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 2.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.3 | 1.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 1.6 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.3 | 2.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 6.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 3.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 4.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.3 | 2.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 2.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 2.5 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 1.9 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.3 | 4.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 1.0 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.3 | 3.8 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 2.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 6.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 3.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.2 | 7.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 1.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 5.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 5.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 6.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 25.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.0 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 3.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 0.8 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.2 | 0.7 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.2 | 4.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 1.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 2.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 1.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 4.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 4.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 2.6 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 5.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 2.0 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.2 | 1.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 6.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.7 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 2.3 | GO:0019871 | potassium channel inhibitor activity(GO:0019870) sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 5.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 2.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.4 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 2.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.1 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.1 | 2.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.0 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.5 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 2.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 5.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 3.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.7 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.7 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 2.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 11.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.4 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.1 | 2.0 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 4.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 3.0 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 2.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.6 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 4.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.8 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.1 | 12.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 1.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 7.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 4.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 2.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.2 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.4 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 2.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 1.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 7.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 4.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.8 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 2.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 4.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 3.2 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 6.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 2.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.8 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 1.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 1.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 4.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 6.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 10.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 1.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 4.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 2.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 1.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 8.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 6.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 6.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 6.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 8.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 9.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 0.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 2.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 4.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 20.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 3.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 3.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 6.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 18.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 15.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 4.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 5.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 12.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 8.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 5.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 3.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 4.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 6.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 5.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 2.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 3.5 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.2 | 11.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 11.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 5.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 5.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 4.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 3.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 4.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.3 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 3.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 4.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 3.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 7.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 3.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 8.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 3.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |