Project
Illumina Body Map 2
Navigation
Downloads
Results for LHX2
Z-value: 1.02
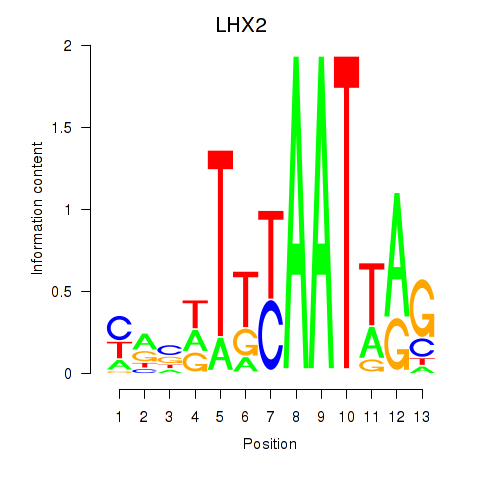
Transcription factors associated with LHX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX2
|
ENSG00000106689.6 | LIM homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX2 | hg19_v2_chr9_+_126773880_126773895 | -0.13 | 4.6e-01 | Click! |
Activity profile of LHX2 motif
Sorted Z-values of LHX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_22554680 | 3.19 |
ENST00000390451.2
|
TRAV23DV6
|
T cell receptor alpha variable 23/delta variable 6 |
| chr9_+_13446472 | 2.67 |
ENST00000428006.2
|
RP11-536O18.1
|
RP11-536O18.1 |
| chr15_-_51535208 | 2.54 |
ENST00000405913.3
ENST00000559878.1 |
CYP19A1
|
cytochrome P450, family 19, subfamily A, polypeptide 1 |
| chr4_-_90756769 | 2.37 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr6_+_26199737 | 2.33 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr13_-_99959641 | 2.23 |
ENST00000376414.4
|
GPR183
|
G protein-coupled receptor 183 |
| chr14_+_22694606 | 2.17 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr14_+_22573582 | 2.05 |
ENST00000390453.1
|
TRAV24
|
T cell receptor alpha variable 24 |
| chr3_+_108541545 | 2.05 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr15_+_51669513 | 1.99 |
ENST00000558426.1
|
GLDN
|
gliomedin |
| chrX_+_99839799 | 1.85 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr7_-_38339890 | 1.82 |
ENST00000390341.2
|
TRGV10
|
T cell receptor gamma variable 10 (non-functional) |
| chr14_+_56584414 | 1.76 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr7_+_80253387 | 1.70 |
ENST00000438020.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr3_-_191000172 | 1.69 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr14_+_23845995 | 1.63 |
ENST00000359320.3
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr13_-_99910673 | 1.61 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr3_+_108541608 | 1.60 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr8_-_82395461 | 1.56 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr14_-_106926724 | 1.54 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr12_+_6554021 | 1.49 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chr2_-_89476644 | 1.48 |
ENST00000484817.1
|
IGKV2-24
|
immunoglobulin kappa variable 2-24 |
| chr5_+_118668846 | 1.48 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr18_+_66465302 | 1.47 |
ENST00000360242.5
ENST00000358653.5 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr2_-_175712270 | 1.43 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr7_+_80254279 | 1.43 |
ENST00000436384.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_+_160709055 | 1.39 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr1_+_160709076 | 1.38 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr4_+_153021899 | 1.37 |
ENST00000509332.1
ENST00000504144.1 ENST00000499452.2 |
RP11-18H21.1
|
RP11-18H21.1 |
| chr4_-_109541539 | 1.36 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr13_-_46952645 | 1.36 |
ENST00000439642.1
|
KIAA0226L
|
KIAA0226-like |
| chr14_+_22992573 | 1.36 |
ENST00000390516.1
|
TRAJ21
|
T cell receptor alpha joining 21 |
| chr1_+_84630574 | 1.33 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr17_-_33864772 | 1.32 |
ENST00000361112.4
|
SLFN12L
|
schlafen family member 12-like |
| chr14_+_22631122 | 1.30 |
ENST00000390458.3
|
TRAV29DV5
|
T cell receptor alpha variable 29/delta variable 5 (gene/pseudogene) |
| chr1_+_81001398 | 1.29 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr11_-_2323089 | 1.28 |
ENST00000456145.2
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr6_+_126661253 | 1.26 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr2_-_175712479 | 1.26 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr16_+_31271274 | 1.24 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr11_-_104972158 | 1.22 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr14_-_107211459 | 1.22 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr3_+_57882061 | 1.20 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr12_+_41136144 | 1.19 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr1_+_160709029 | 1.16 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr16_+_31213206 | 1.15 |
ENST00000561916.2
|
C16orf98
|
chromosome 16 open reading frame 98 |
| chr6_-_26199499 | 1.15 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr13_-_99910620 | 1.14 |
ENST00000416594.1
|
GPR18
|
G protein-coupled receptor 18 |
| chr1_+_192127578 | 1.13 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr2_-_89157161 | 1.11 |
ENST00000390237.2
|
IGKC
|
immunoglobulin kappa constant |
| chr14_-_78083112 | 1.10 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr15_-_72521017 | 1.09 |
ENST00000561609.1
|
PKM
|
pyruvate kinase, muscle |
| chr14_+_52313833 | 1.08 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr2_+_87769459 | 1.07 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr4_+_70796784 | 1.07 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr17_-_295730 | 1.05 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr3_+_57881966 | 1.03 |
ENST00000495364.1
|
SLMAP
|
sarcolemma associated protein |
| chr2_-_112237835 | 1.02 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr11_-_40315640 | 1.01 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr11_-_46113756 | 1.01 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr6_-_26199471 | 1.00 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr5_-_41794313 | 0.99 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr4_-_120243545 | 0.99 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr2_+_90139056 | 0.96 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr13_-_19755975 | 0.94 |
ENST00000400113.3
|
TUBA3C
|
tubulin, alpha 3c |
| chr2_+_68961934 | 0.93 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr1_-_153518270 | 0.92 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr8_-_19459993 | 0.91 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr7_-_38315919 | 0.90 |
ENST00000390339.1
|
TRGJP1
|
T cell receptor gamma joining P1 |
| chr2_+_68961905 | 0.90 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr17_-_57229155 | 0.88 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr7_-_142232071 | 0.88 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr1_-_183560011 | 0.87 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr2_+_68962014 | 0.85 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr4_+_70916119 | 0.84 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr3_+_57882024 | 0.84 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr19_+_20188830 | 0.83 |
ENST00000418063.2
|
ZNF90
|
zinc finger protein 90 |
| chr14_+_22564294 | 0.83 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1 |
| chr12_+_9980069 | 0.82 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr2_-_231084617 | 0.81 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr1_-_168464875 | 0.80 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr8_-_109260897 | 0.75 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chrX_-_11129229 | 0.75 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr2_-_231084659 | 0.75 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr2_-_235363231 | 0.74 |
ENST00000439798.1
|
AC097713.4
|
AC097713.4 |
| chr4_-_159956333 | 0.73 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr12_+_48722763 | 0.73 |
ENST00000335017.1
|
H1FNT
|
H1 histone family, member N, testis-specific |
| chr20_-_56647116 | 0.73 |
ENST00000441277.2
ENST00000452842.1 |
RP13-379L11.1
|
RP13-379L11.1 |
| chr2_+_162480845 | 0.72 |
ENST00000375514.5
ENST00000415876.2 |
SLC4A10
|
solute carrier family 4, sodium bicarbonate transporter, member 10 |
| chr1_-_152332480 | 0.70 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
| chr11_-_132812987 | 0.70 |
ENST00000541867.1
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr15_-_33180439 | 0.69 |
ENST00000559610.1
|
FMN1
|
formin 1 |
| chr10_+_4828815 | 0.68 |
ENST00000533295.1
|
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr14_+_23002427 | 0.68 |
ENST00000390527.1
|
TRAJ10
|
T cell receptor alpha joining 10 |
| chr2_+_74648848 | 0.68 |
ENST00000409791.1
ENST00000426787.1 ENST00000348227.4 |
WDR54
|
WD repeat domain 54 |
| chr1_+_244515930 | 0.67 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr7_-_14880892 | 0.66 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr9_+_105757590 | 0.64 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr2_+_200472779 | 0.63 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr10_-_4285923 | 0.62 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr14_-_22005343 | 0.61 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chrX_+_18725758 | 0.61 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr8_+_66955648 | 0.61 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr4_-_153332886 | 0.59 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr14_+_100532771 | 0.59 |
ENST00000557153.1
|
EVL
|
Enah/Vasp-like |
| chr16_+_24266874 | 0.59 |
ENST00000005284.3
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr18_+_32556892 | 0.59 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr4_+_69313145 | 0.57 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr13_+_24844979 | 0.57 |
ENST00000454083.1
|
SPATA13
|
spermatogenesis associated 13 |
| chrY_+_15418467 | 0.57 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr16_+_333152 | 0.56 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr2_+_210517895 | 0.54 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr13_+_24844857 | 0.54 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr3_+_62936098 | 0.54 |
ENST00000475886.1
ENST00000465684.1 ENST00000465262.1 ENST00000468072.1 |
LINC00698
|
long intergenic non-protein coding RNA 698 |
| chr7_-_5998714 | 0.52 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr2_-_238343455 | 0.52 |
ENST00000409910.1
|
AC112721.2
|
AC112721.2 |
| chr19_+_42212526 | 0.52 |
ENST00000598976.1
ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA
CEACAM5
|
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr5_+_180682720 | 0.52 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr11_-_62323702 | 0.51 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr5_-_75008244 | 0.51 |
ENST00000510798.1
ENST00000446329.2 |
POC5
|
POC5 centriolar protein |
| chr9_-_139137648 | 0.51 |
ENST00000358701.5
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr4_+_48485341 | 0.51 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chr5_+_179921344 | 0.51 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr3_+_33155444 | 0.50 |
ENST00000320954.6
|
CRTAP
|
cartilage associated protein |
| chr4_+_70894130 | 0.49 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr13_-_51101468 | 0.47 |
ENST00000428276.1
|
RP11-175B12.2
|
RP11-175B12.2 |
| chr11_-_104827425 | 0.47 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr17_+_61086917 | 0.46 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr6_+_130339710 | 0.46 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chrX_-_11284095 | 0.46 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr11_-_108093329 | 0.45 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr8_-_37411648 | 0.44 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr7_-_65113280 | 0.44 |
ENST00000593865.1
|
AC104057.1
|
Protein LOC100996407 |
| chr5_+_137722255 | 0.44 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr3_+_73110810 | 0.42 |
ENST00000533473.1
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2 |
| chr6_-_131291572 | 0.42 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr14_-_34420259 | 0.42 |
ENST00000250457.3
ENST00000547327.2 |
EGLN3
|
egl-9 family hypoxia-inducible factor 3 |
| chr2_-_99279928 | 0.40 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr2_-_37501692 | 0.40 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr12_-_25348007 | 0.39 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr12_-_118796971 | 0.38 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr19_+_42212501 | 0.37 |
ENST00000398599.4
|
CEACAM5
|
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr13_+_33160553 | 0.36 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr13_+_24844819 | 0.36 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr9_-_28670283 | 0.35 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr8_-_93029865 | 0.35 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_-_28527152 | 0.35 |
ENST00000321830.5
|
AL353354.1
|
Uncharacterized protein |
| chr18_+_34124507 | 0.35 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr12_-_49245916 | 0.35 |
ENST00000552512.1
ENST00000551468.1 |
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr3_+_119187785 | 0.34 |
ENST00000295588.4
ENST00000476573.1 |
POGLUT1
|
protein O-glucosyltransferase 1 |
| chr15_-_42565023 | 0.34 |
ENST00000566474.1
|
TMEM87A
|
transmembrane protein 87A |
| chr1_-_232651312 | 0.33 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr18_+_3247779 | 0.33 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr19_+_22235310 | 0.32 |
ENST00000600162.1
|
ZNF257
|
zinc finger protein 257 |
| chr7_-_22234381 | 0.32 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr12_-_118796910 | 0.32 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr10_-_116286656 | 0.31 |
ENST00000428430.1
ENST00000369266.3 ENST00000392952.3 |
ABLIM1
|
actin binding LIM protein 1 |
| chr10_-_90712520 | 0.31 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr2_+_29001711 | 0.31 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr13_+_78315528 | 0.30 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr13_+_19756173 | 0.30 |
ENST00000382988.2
|
RP11-408E5.4
|
RP11-408E5.4 |
| chr7_+_101460882 | 0.30 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr5_+_68860949 | 0.30 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr17_+_48823975 | 0.29 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr14_+_22251210 | 0.29 |
ENST00000390429.3
|
TRAV7
|
T cell receptor alpha variable 7 |
| chr1_+_87012922 | 0.29 |
ENST00000263723.5
|
CLCA4
|
chloride channel accessory 4 |
| chr11_-_55371874 | 0.28 |
ENST00000302231.4
|
OR4C11
|
olfactory receptor, family 4, subfamily C, member 11 |
| chr4_+_147096837 | 0.28 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr7_+_99717230 | 0.27 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr5_-_150473127 | 0.27 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr19_-_44384291 | 0.27 |
ENST00000324394.6
|
ZNF404
|
zinc finger protein 404 |
| chr18_+_3466248 | 0.27 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr4_+_128554081 | 0.27 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr18_+_29027696 | 0.27 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr14_+_29299437 | 0.26 |
ENST00000550827.1
ENST00000548087.1 ENST00000551588.1 ENST00000550122.1 |
CTD-2384A14.1
|
CTD-2384A14.1 |
| chr11_+_24518723 | 0.26 |
ENST00000336930.6
ENST00000529015.1 ENST00000533227.1 |
LUZP2
|
leucine zipper protein 2 |
| chr20_+_31870927 | 0.26 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr15_+_54793169 | 0.25 |
ENST00000559093.1
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr7_-_15014398 | 0.25 |
ENST00000437998.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr21_-_43735446 | 0.25 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr8_+_92114060 | 0.24 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr5_+_98109322 | 0.24 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr9_-_98268883 | 0.24 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chrX_-_19988382 | 0.23 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr12_-_118797475 | 0.23 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr13_+_77522632 | 0.22 |
ENST00000377462.1
|
IRG1
|
immunoresponsive 1 homolog (mouse) |
| chr1_-_75100539 | 0.22 |
ENST00000420661.2
|
C1orf173
|
chromosome 1 open reading frame 173 |
| chr15_-_72563585 | 0.22 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr7_+_156433573 | 0.20 |
ENST00000311822.8
|
RNF32
|
ring finger protein 32 |
| chr18_-_19994830 | 0.20 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr9_+_116207007 | 0.19 |
ENST00000374140.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr3_+_98001732 | 0.18 |
ENST00000355273.2
|
OR5H2
|
olfactory receptor, family 5, subfamily H, member 2 |
| chr8_-_82373809 | 0.18 |
ENST00000379071.2
|
FABP9
|
fatty acid binding protein 9, testis |
| chr16_+_23652773 | 0.18 |
ENST00000563998.1
ENST00000568589.1 ENST00000568272.1 |
DCTN5
|
dynactin 5 (p25) |
| chr12_+_6309517 | 0.18 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr17_+_17082842 | 0.18 |
ENST00000579361.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr1_-_89591749 | 0.17 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr7_-_99716914 | 0.17 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr10_-_24770632 | 0.17 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr13_+_78315466 | 0.17 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0035700 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.7 | 2.8 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.6 | 2.5 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.5 | 2.4 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.4 | 3.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 3.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.4 | 1.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.3 | 1.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.3 | 1.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.3 | 1.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.3 | 1.8 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.3 | 3.1 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 0.5 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.1 | 1.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 1.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.6 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.9 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 1.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.7 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 2.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 0.7 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 1.6 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 1.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.8 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 1.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.3 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 3.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 2.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 1.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 1.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 1.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 3.7 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 1.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.9 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 1.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.7 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0060754 | regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.7 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 1.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.5 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 2.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.5 | GO:0007049 | cell cycle(GO:0007049) |
| 0.0 | 0.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.2 | 0.6 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.2 | 1.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 1.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 5.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.7 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 3.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.9 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 1.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 3.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 2.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.9 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.0 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.7 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 3.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 1.0 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 0.7 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.2 | 2.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.9 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 3.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.5 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 1.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 1.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.9 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 2.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.8 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 3.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 2.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 2.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 4.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 3.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 4.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 3.6 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 2.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 4.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |