Project
Illumina Body Map 2
Navigation
Downloads
Results for LHX8
Z-value: 0.79
Transcription factors associated with LHX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX8
|
ENSG00000162624.10 | LIM homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX8 | hg19_v2_chr1_+_75594119_75594119 | -0.27 | 1.4e-01 | Click! |
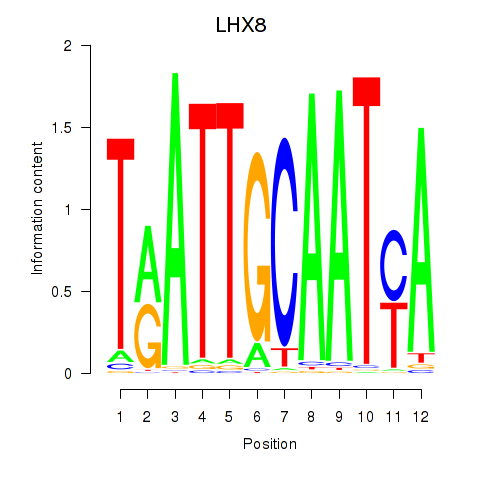
Activity profile of LHX8 motif
Sorted Z-values of LHX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_54982420 | 2.32 |
ENST00000257905.8
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr1_+_145413268 | 2.27 |
ENST00000421822.2
ENST00000336751.5 ENST00000497365.1 ENST00000475797.1 |
HFE2
|
hemochromatosis type 2 (juvenile) |
| chr2_+_168043793 | 1.58 |
ENST00000409273.1
ENST00000409605.1 |
XIRP2
|
xin actin-binding repeat containing 2 |
| chr10_+_69869237 | 1.42 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr12_-_54982300 | 1.36 |
ENST00000547431.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr7_-_120498357 | 1.34 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr12_+_101988627 | 1.28 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr8_+_9009296 | 1.15 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr12_+_101988774 | 1.14 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr2_-_183291741 | 1.11 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chrX_-_21776281 | 0.96 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chrX_-_73051037 | 0.93 |
ENST00000445814.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr4_+_74275057 | 0.90 |
ENST00000511370.1
|
ALB
|
albumin |
| chr8_-_82608409 | 0.89 |
ENST00000518568.1
|
SLC10A5
|
solute carrier family 10, member 5 |
| chr8_-_25902876 | 0.87 |
ENST00000520164.1
|
EBF2
|
early B-cell factor 2 |
| chr4_-_100242549 | 0.86 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr5_+_140235469 | 0.84 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr8_-_82395461 | 0.84 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr19_-_4535233 | 0.84 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr2_+_234621551 | 0.83 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr8_+_19759228 | 0.82 |
ENST00000520959.1
|
LPL
|
lipoprotein lipase |
| chr7_-_137028534 | 0.80 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr2_-_225266802 | 0.79 |
ENST00000243806.2
|
FAM124B
|
family with sequence similarity 124B |
| chr21_+_39644305 | 0.79 |
ENST00000398930.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr4_+_166300084 | 0.76 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr7_-_137028498 | 0.74 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr19_-_58204128 | 0.72 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr12_+_48516357 | 0.70 |
ENST00000549022.1
ENST00000547587.1 ENST00000312352.7 |
PFKM
|
phosphofructokinase, muscle |
| chr4_-_69111401 | 0.70 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr15_-_34880646 | 0.70 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr6_-_123958141 | 0.69 |
ENST00000334268.4
|
TRDN
|
triadin |
| chr4_-_186734275 | 0.69 |
ENST00000456060.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_+_84258832 | 0.69 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr1_+_47603109 | 0.68 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr1_-_217262933 | 0.67 |
ENST00000359162.2
|
ESRRG
|
estrogen-related receptor gamma |
| chr6_-_24489842 | 0.65 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr18_+_71815743 | 0.65 |
ENST00000169551.6
ENST00000580087.1 |
TIMM21
|
translocase of inner mitochondrial membrane 21 homolog (yeast) |
| chr10_+_24738355 | 0.64 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr11_-_117800080 | 0.63 |
ENST00000524993.1
ENST00000528626.1 ENST00000445164.2 ENST00000430170.2 ENST00000526090.1 |
TMPRSS13
|
transmembrane protease, serine 13 |
| chrX_-_5644225 | 0.63 |
ENST00000422914.1
|
RP11-733O18.1
|
RP11-733O18.1 |
| chr5_+_140593509 | 0.61 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chrX_+_70798261 | 0.61 |
ENST00000373696.3
|
ACRC
|
acidic repeat containing |
| chr13_+_24883703 | 0.61 |
ENST00000332018.4
|
C1QTNF9
|
C1q and tumor necrosis factor related protein 9 |
| chr14_-_36990354 | 0.61 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr1_+_95616933 | 0.61 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr12_+_48516463 | 0.60 |
ENST00000546465.1
|
PFKM
|
phosphofructokinase, muscle |
| chr1_-_217262969 | 0.60 |
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr4_-_186733363 | 0.60 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_110723134 | 0.58 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr5_+_135170331 | 0.57 |
ENST00000425402.1
ENST00000274513.5 ENST00000420621.1 ENST00000433282.2 ENST00000412661.2 |
SLC25A48
|
solute carrier family 25, member 48 |
| chr3_+_154798162 | 0.57 |
ENST00000360490.2
|
MME
|
membrane metallo-endopeptidase |
| chr1_+_70876926 | 0.56 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr4_+_155484103 | 0.56 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr22_+_22516550 | 0.53 |
ENST00000390284.2
|
IGLV4-60
|
immunoglobulin lambda variable 4-60 |
| chr8_+_101349823 | 0.53 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr6_+_153071925 | 0.53 |
ENST00000367244.3
ENST00000367243.3 |
VIP
|
vasoactive intestinal peptide |
| chr6_-_56507586 | 0.53 |
ENST00000439203.1
ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST
|
dystonin |
| chr21_-_34863998 | 0.53 |
ENST00000402202.1
ENST00000381947.3 |
DNAJC28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr18_-_11908272 | 0.52 |
ENST00000592977.1
ENST00000590501.1 ENST00000586844.1 |
MPPE1
|
metallophosphoesterase 1 |
| chr2_+_201994208 | 0.52 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_102758210 | 0.51 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr4_-_110723335 | 0.51 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr4_-_110723194 | 0.50 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr9_-_70490107 | 0.49 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr11_+_60048004 | 0.49 |
ENST00000532114.1
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr1_+_144989309 | 0.49 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr12_-_15038779 | 0.49 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr18_-_11908329 | 0.48 |
ENST00000344987.7
ENST00000588103.1 ENST00000588191.1 ENST00000317235.7 ENST00000309976.9 ENST00000588186.1 ENST00000589267.1 |
MPPE1
|
metallophosphoesterase 1 |
| chr11_+_60048129 | 0.47 |
ENST00000355131.3
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr2_+_102758271 | 0.47 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr9_+_70856899 | 0.47 |
ENST00000377342.5
ENST00000478048.1 |
CBWD3
|
COBW domain containing 3 |
| chr6_-_44400720 | 0.46 |
ENST00000595057.1
|
AL133262.1
|
AL133262.1 |
| chr7_+_151791095 | 0.46 |
ENST00000422997.2
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr10_+_81892347 | 0.45 |
ENST00000372267.2
|
PLAC9
|
placenta-specific 9 |
| chr9_-_13165457 | 0.45 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr1_+_53662101 | 0.45 |
ENST00000371486.3
|
CPT2
|
carnitine palmitoyltransferase 2 |
| chr6_+_143759575 | 0.45 |
ENST00000595616.1
|
AL031320.1
|
AL031320.1 |
| chr9_-_69262509 | 0.45 |
ENST00000377449.1
ENST00000382399.4 ENST00000377439.1 ENST00000377441.1 ENST00000377457.5 |
CBWD6
|
COBW domain containing 6 |
| chr1_+_230138476 | 0.45 |
ENST00000445339.1
|
RP11-552D4.1
|
RP11-552D4.1 |
| chr21_-_16254231 | 0.44 |
ENST00000412426.1
ENST00000418954.1 |
AF127936.7
|
AF127936.7 |
| chr2_+_114195268 | 0.44 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr6_-_123958111 | 0.44 |
ENST00000542443.1
|
TRDN
|
triadin |
| chr19_+_12175504 | 0.43 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr8_-_62602327 | 0.43 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr14_+_57046500 | 0.43 |
ENST00000261556.6
|
TMEM260
|
transmembrane protein 260 |
| chr2_+_201994042 | 0.43 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr8_-_30013748 | 0.43 |
ENST00000607315.1
|
RP11-51J9.5
|
RP11-51J9.5 |
| chr1_-_212004090 | 0.43 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr17_+_58018269 | 0.43 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr7_+_114055052 | 0.42 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr12_-_14967095 | 0.42 |
ENST00000316048.2
|
SMCO3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr2_-_152830441 | 0.42 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr1_-_168464875 | 0.42 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr9_-_179018 | 0.41 |
ENST00000431099.2
ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1
|
COBW domain containing 1 |
| chr12_+_59989918 | 0.41 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_+_108145913 | 0.41 |
ENST00000443205.1
|
AC096669.3
|
AC096669.3 |
| chr2_-_14541060 | 0.41 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr9_+_70856397 | 0.41 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr22_+_23487513 | 0.40 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr16_-_20338748 | 0.40 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr9_-_20622478 | 0.40 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr9_-_5833027 | 0.40 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr3_+_195447738 | 0.40 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chrX_-_131262048 | 0.39 |
ENST00000298542.4
|
FRMD7
|
FERM domain containing 7 |
| chr6_+_32006042 | 0.39 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr4_-_109684120 | 0.39 |
ENST00000512646.1
ENST00000411864.2 ENST00000296486.3 ENST00000510706.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr14_+_57046530 | 0.39 |
ENST00000536419.1
ENST00000538838.1 |
TMEM260
|
transmembrane protein 260 |
| chr4_+_155484155 | 0.38 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr8_-_112039643 | 0.38 |
ENST00000524283.1
|
RP11-946L20.2
|
RP11-946L20.2 |
| chr3_-_151102529 | 0.38 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr6_-_4347271 | 0.36 |
ENST00000437430.2
|
RP3-527G5.1
|
RP3-527G5.1 |
| chr7_+_141408153 | 0.36 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr3_+_173116225 | 0.36 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr6_+_26087646 | 0.35 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr2_-_152830479 | 0.35 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr6_+_26087509 | 0.35 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr6_-_75828774 | 0.35 |
ENST00000493109.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr7_+_7811992 | 0.34 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr7_+_151791074 | 0.33 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr6_+_76599809 | 0.33 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr11_+_1049862 | 0.33 |
ENST00000534584.1
|
RP13-870H17.3
|
RP13-870H17.3 |
| chr2_-_136678123 | 0.33 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr16_-_20339123 | 0.32 |
ENST00000381360.5
|
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr11_-_115127611 | 0.32 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr11_+_72975524 | 0.32 |
ENST00000540342.1
ENST00000542092.1 |
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr16_+_22217577 | 0.31 |
ENST00000263026.5
|
EEF2K
|
eukaryotic elongation factor-2 kinase |
| chr20_+_56964169 | 0.31 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr15_+_86805875 | 0.31 |
ENST00000389298.3
|
AGBL1
|
ATP/GTP binding protein-like 1 |
| chr15_+_65843130 | 0.30 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr2_-_225266711 | 0.30 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124B |
| chr7_-_92777606 | 0.30 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr8_-_141810634 | 0.30 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr10_+_75668916 | 0.30 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr12_-_88974236 | 0.29 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr2_-_225266743 | 0.28 |
ENST00000409685.3
|
FAM124B
|
family with sequence similarity 124B |
| chrX_+_73164167 | 0.28 |
ENST00000414209.1
ENST00000602895.1 ENST00000453317.1 ENST00000602546.1 ENST00000602985.1 ENST00000415215.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr8_+_17354617 | 0.28 |
ENST00000470360.1
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr6_-_152623231 | 0.27 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr9_+_108424738 | 0.27 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr19_+_34856078 | 0.27 |
ENST00000589399.1
ENST00000589640.1 ENST00000591204.1 |
GPI
|
glucose-6-phosphate isomerase |
| chr12_-_10766184 | 0.27 |
ENST00000539554.1
ENST00000381881.2 ENST00000320756.2 |
MAGOHB
|
mago-nashi homolog B (Drosophila) |
| chr10_-_110321541 | 0.27 |
ENST00000366253.2
|
RP11-163F15.1
|
RP11-163F15.1 |
| chr5_-_41213607 | 0.27 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr11_+_60048053 | 0.27 |
ENST00000337908.4
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr12_+_122326630 | 0.26 |
ENST00000541212.1
ENST00000340175.5 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr7_+_30589829 | 0.26 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr14_+_50291993 | 0.26 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr6_+_10633993 | 0.26 |
ENST00000417671.1
|
GCNT6
|
glucosaminyl (N-acetyl) transferase 6 |
| chr13_-_24895566 | 0.26 |
ENST00000422229.2
|
AL359736.1
|
protein PCOTH isoform 1 |
| chr15_+_54793169 | 0.26 |
ENST00000559093.1
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr5_+_140165876 | 0.26 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr11_-_3400442 | 0.26 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr11_-_58035732 | 0.25 |
ENST00000395079.2
|
OR10W1
|
olfactory receptor, family 10, subfamily W, member 1 |
| chr13_+_38923959 | 0.25 |
ENST00000379649.1
ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1
|
ubiquitin-fold modifier 1 |
| chr8_+_39806685 | 0.25 |
ENST00000389060.4
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chrX_+_73164149 | 0.25 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr12_-_39734783 | 0.25 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr1_+_66796401 | 0.25 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr3_-_150612089 | 0.25 |
ENST00000397891.3
|
FAM188B2
|
family with sequence similarity 188, member B2 |
| chrX_-_23926004 | 0.25 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr2_+_216946589 | 0.25 |
ENST00000433112.1
ENST00000454545.1 ENST00000437356.2 ENST00000295658.4 ENST00000455479.1 ENST00000406027.2 |
TMEM169
|
transmembrane protein 169 |
| chr1_-_95391315 | 0.25 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr11_+_72975578 | 0.25 |
ENST00000393592.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr3_-_47934234 | 0.24 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr18_-_74839891 | 0.24 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr4_-_69083720 | 0.24 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr3_-_87842631 | 0.24 |
ENST00000462792.1
|
RP11-451B8.1
|
RP11-451B8.1 |
| chr1_+_145883868 | 0.24 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr3_+_148545586 | 0.24 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr1_-_110933611 | 0.24 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr1_-_110933663 | 0.23 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr2_+_86947296 | 0.23 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr19_+_12175535 | 0.23 |
ENST00000550826.1
|
ZNF844
|
zinc finger protein 844 |
| chr8_+_119294456 | 0.23 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr2_-_37068530 | 0.23 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr7_-_22234381 | 0.23 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr12_+_26348246 | 0.22 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr2_+_234826016 | 0.22 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr19_+_42037433 | 0.22 |
ENST00000599316.1
ENST00000599770.1 |
AC006129.1
|
AC006129.1 |
| chr6_+_43543942 | 0.22 |
ENST00000372226.1
ENST00000443535.1 |
POLH
|
polymerase (DNA directed), eta |
| chr7_-_20256965 | 0.22 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr7_+_151791037 | 0.21 |
ENST00000419245.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr5_+_140480083 | 0.21 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr5_+_147715119 | 0.21 |
ENST00000377906.1
|
SPINK9
|
serine peptidase inhibitor, Kazal type 9 |
| chr9_+_96846740 | 0.20 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr19_+_34855925 | 0.20 |
ENST00000590375.1
ENST00000356487.5 |
GPI
|
glucose-6-phosphate isomerase |
| chr8_+_17354587 | 0.20 |
ENST00000494857.1
ENST00000522656.1 |
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr17_-_3301704 | 0.19 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1 |
| chr11_-_104817919 | 0.19 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr10_+_135207598 | 0.19 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr19_+_34855874 | 0.19 |
ENST00000588991.2
|
GPI
|
glucose-6-phosphate isomerase |
| chr3_-_187455680 | 0.19 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr19_+_21688366 | 0.19 |
ENST00000358491.4
ENST00000597078.1 |
ZNF429
|
zinc finger protein 429 |
| chr7_-_7575477 | 0.19 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr19_+_21264980 | 0.19 |
ENST00000596053.1
ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714
|
zinc finger protein 714 |
| chr11_+_72975559 | 0.19 |
ENST00000349767.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chrX_+_56258844 | 0.18 |
ENST00000374928.3
|
KLF8
|
Kruppel-like factor 8 |
| chr1_-_146040968 | 0.17 |
ENST00000401010.3
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr1_-_247242048 | 0.17 |
ENST00000366503.2
|
ZNF670
|
zinc finger protein 670 |
| chr5_-_150473127 | 0.17 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.3 | 0.9 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.3 | 1.1 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 0.3 | 0.8 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 0.7 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.2 | 1.3 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.2 | 0.5 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.2 | 0.8 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.2 | 1.0 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.2 | 0.6 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.2 | 0.5 | GO:0034226 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.1 | 0.6 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.1 | 0.4 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 2.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.4 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.3 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.7 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.1 | 0.3 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 2.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.4 | GO:0097115 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.9 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.0 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 1.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.6 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.1 | 0.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.3 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.6 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.4 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.6 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.4 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.8 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.5 | GO:0070459 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) prolactin secretion(GO:0070459) |
| 0.0 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.9 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 1.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 1.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 3.7 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 0.1 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.3 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0035587 | purinergic receptor signaling pathway(GO:0035587) purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 1.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.0 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.3 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0043504 | DNA replication, removal of RNA primer(GO:0043137) mitochondrial DNA repair(GO:0043504) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 1.6 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.0 | GO:1901963 | glial cell fate determination(GO:0007403) oviduct development(GO:0060066) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.0 | GO:1902304 | phosphatidylserine exposure on blood platelet(GO:0097045) positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.0 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.6 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 1.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 2.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.9 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 1.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 1.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.0 | GO:0000793 | chromosome, centromeric region(GO:0000775) condensed chromosome(GO:0000793) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 2.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.3 | 2.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 3.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 0.8 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 1.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 1.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.4 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 1.0 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.7 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.1 | 1.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.3 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.1 | 0.8 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 3.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.4 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.9 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 1.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.9 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.7 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 1.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.6 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.8 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 1.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.4 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 2.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 2.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 2.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |