Project
Illumina Body Map 2
Navigation
Downloads
Results for MLX_USF2_USF1_PAX2
Z-value: 1.32
Transcription factors associated with MLX_USF2_USF1_PAX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
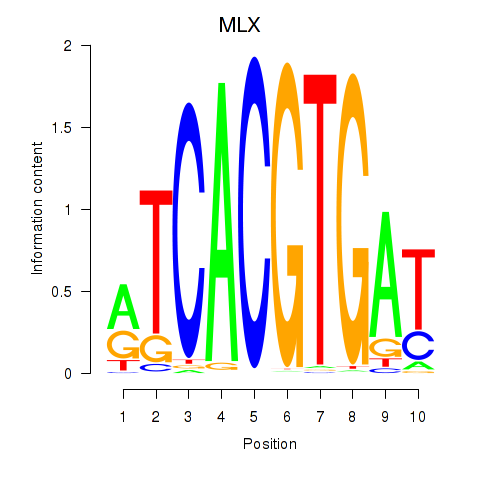
MLX
|
ENSG00000108788.7 | MAX dimerization protein MLX |
|
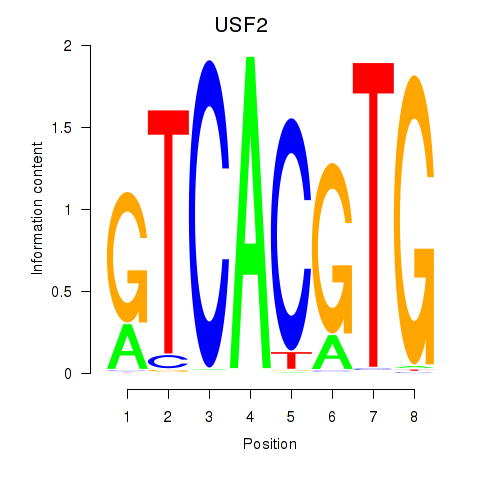
USF2
|
ENSG00000105698.11 | upstream transcription factor 2, c-fos interacting |
|
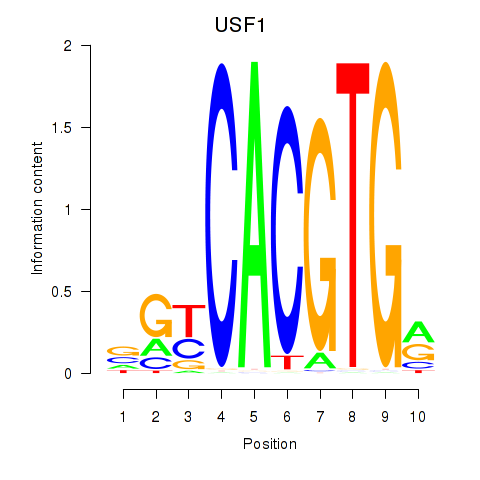
USF1
|
ENSG00000158773.10 | upstream transcription factor 1 |
|
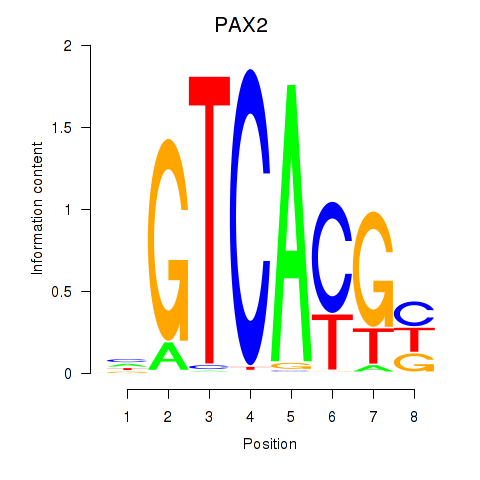
PAX2
|
ENSG00000075891.17 | paired box 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MLX | hg19_v2_chr17_+_40719073_40719092 | 0.39 | 2.9e-02 | Click! |
| USF1 | hg19_v2_chr1_-_161015752_161015778 | 0.35 | 4.6e-02 | Click! |
| USF2 | hg19_v2_chr19_+_35759824_35759891 | 0.20 | 2.7e-01 | Click! |
| PAX2 | hg19_v2_chr10_+_102505468_102505546 | -0.14 | 4.6e-01 | Click! |
Activity profile of MLX_USF2_USF1_PAX2 motif
Sorted Z-values of MLX_USF2_USF1_PAX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MLX_USF2_USF1_PAX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 14.3 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 1.6 | 6.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 1.4 | 5.7 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 1.4 | 9.5 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 1.3 | 5.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 1.3 | 1.3 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 1.2 | 4.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 1.1 | 4.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 1.0 | 2.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 1.0 | 4.9 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 1.0 | 4.8 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.9 | 5.4 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.8 | 10.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.8 | 3.3 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.8 | 2.4 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.7 | 2.8 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.7 | 2.0 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.7 | 2.0 | GO:0070429 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.6 | 5.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.6 | 2.5 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.6 | 1.8 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.6 | 2.3 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.6 | 4.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.5 | 4.9 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.5 | 9.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.5 | 5.8 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.5 | 1.5 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.5 | 1.5 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.5 | 3.4 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.5 | 6.8 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.5 | 1.4 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.5 | 1.8 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.5 | 1.8 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.4 | 2.2 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.4 | 0.9 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.4 | 3.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.4 | 2.6 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 1.3 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.4 | 4.7 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.4 | 0.4 | GO:0043318 | regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.4 | 1.2 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.4 | 2.0 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.4 | 1.6 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.4 | 2.0 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 1.6 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.4 | 4.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.4 | 1.5 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.4 | 3.4 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.4 | 2.2 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.4 | 1.5 | GO:0090678 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.4 | 1.1 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.4 | 1.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.4 | 3.6 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.4 | 3.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.4 | 1.4 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.4 | 3.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.3 | 5.2 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.3 | 1.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.3 | 2.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.3 | 2.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 1.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.3 | 7.0 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.3 | 0.9 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.3 | 0.3 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.3 | 2.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 1.8 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.3 | 0.9 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.3 | 0.9 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 0.3 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.3 | 11.3 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.3 | 0.9 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.3 | 1.5 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.3 | 5.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.3 | 2.9 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.3 | 1.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.3 | 1.7 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.3 | 0.3 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.3 | 0.8 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.3 | 0.8 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.3 | 5.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 0.3 | GO:2000506 | negative regulation of energy homeostasis(GO:2000506) |
| 0.3 | 0.3 | GO:0071506 | response to mycophenolic acid(GO:0071505) cellular response to mycophenolic acid(GO:0071506) |
| 0.3 | 3.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.3 | 2.4 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.3 | 1.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.3 | 1.3 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.3 | 1.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 0.8 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.3 | 0.8 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 1.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.3 | 1.3 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.3 | 3.0 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.3 | 1.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.2 | 0.7 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 0.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 1.2 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.2 | 2.2 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.2 | 2.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 0.7 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.2 | 0.7 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.2 | 0.5 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 | 0.7 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 1.6 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.2 | 2.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.2 | 2.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 0.9 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.1 | GO:0039650 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.2 | 1.5 | GO:2000771 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.2 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.2 | 0.8 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.2 | 1.9 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 0.8 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 1.0 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 1.0 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.2 | 2.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 3.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 1.0 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 0.8 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.2 | 0.8 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 0.8 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.2 | 0.8 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.2 | 0.6 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 3.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.2 | 0.6 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 | 0.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.5 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.2 | 0.7 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.2 | 3.4 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.2 | 0.5 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 3.0 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 0.5 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 0.9 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.2 | 0.7 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.2 | 1.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 2.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 0.2 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.2 | 2.9 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.2 | 1.0 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 | 0.8 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 | 1.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 0.5 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.2 | 0.5 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.2 | 1.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.2 | 0.8 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.2 | 2.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 1.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.4 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 1.5 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 1.2 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 5.0 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.7 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 3.7 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.4 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.7 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 1.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 2.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 1.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.5 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 2.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 3.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.7 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 3.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.4 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.1 | 2.1 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.1 | 0.4 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.4 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.9 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 5.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.7 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.1 | 0.5 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.4 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 0.6 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 6.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.8 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.3 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 1.2 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.1 | 0.3 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.5 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 1.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 2.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.7 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.6 | GO:1904884 | establishment of RNA localization to telomere(GO:0097694) establishment of macromolecular complex localization to telomere(GO:0097695) positive regulation of DNA catabolic process(GO:1903626) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 1.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 2.9 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.3 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.1 | 0.2 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 1.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.1 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.1 | 0.6 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.2 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 1.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.5 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.6 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.1 | 0.4 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 1.7 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.3 | GO:0042921 | corticosteroid receptor signaling pathway(GO:0031958) glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 0.5 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 1.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.7 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.1 | 1.7 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 1.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.5 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.9 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 1.9 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 6.1 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.1 | 0.6 | GO:0006549 | allantoin metabolic process(GO:0000255) isoleucine metabolic process(GO:0006549) |
| 0.1 | 1.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.2 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 1.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.7 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.4 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.3 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.3 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 1.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.5 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 5.7 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 1.0 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.2 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.8 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.4 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 1.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 1.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 2.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.5 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.1 | 0.8 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.7 | GO:0035898 | parathyroid hormone secretion(GO:0035898) post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.1 | 0.7 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.1 | 0.7 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.1 | 0.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.7 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 1.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.6 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 0.3 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.3 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.1 | 1.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 1.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 0.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.8 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 1.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.3 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.1 | 0.3 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 1.9 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 0.3 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.5 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.4 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.5 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 4.8 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.1 | 0.5 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.1 | 0.6 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.8 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.4 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.4 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.5 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 4.9 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 0.9 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.2 | GO:0035349 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.1 | 0.5 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 0.2 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.6 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.2 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 0.8 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 0.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.5 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 2.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 2.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 2.9 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.1 | 0.4 | GO:0071362 | cellular response to ether(GO:0071362) |
| 0.1 | 2.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 2.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.2 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.4 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 0.4 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 1.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.9 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.5 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 0.9 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.1 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.4 | GO:0034030 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.3 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 0.2 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 1.5 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 | 0.7 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.7 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.6 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 0.2 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 0.3 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.1 | 1.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 8.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 1.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.2 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.2 | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.1 | 1.7 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.1 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.9 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 0.8 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 1.2 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.2 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 0.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 1.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 2.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.9 | GO:0000075 | cell cycle checkpoint(GO:0000075) |
| 0.1 | 1.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.2 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.1 | 2.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 3.8 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 1.7 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 2.9 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 0.4 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 0.2 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.5 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.7 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.4 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.8 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.5 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.2 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.3 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.3 | GO:0006014 | D-ribose metabolic process(GO:0006014) pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.8 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 1.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.6 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.7 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.0 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.5 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 1.1 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.8 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.3 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 2.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.5 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.7 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.9 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:2000641 | regulation of early endosome to late endosome transport(GO:2000641) |
| 0.0 | 2.3 | GO:0006735 | NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.6 | GO:0070572 | positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.4 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.5 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 2.8 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.3 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 1.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.8 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.4 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 0.6 | GO:0046415 | urate transport(GO:0015747) urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 1.4 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.0 | 3.5 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.5 | GO:0014894 | response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 2.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.4 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.2 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 3.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 2.0 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.2 | GO:1902741 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 2.0 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.7 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0060754 | regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.7 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 1.5 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.8 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.5 | GO:0071402 | cellular response to lipoprotein particle stimulus(GO:0071402) |
| 0.0 | 0.1 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 1.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0010452 | regulation of histone H3-K36 methylation(GO:0000414) histone H3-K36 methylation(GO:0010452) |
| 0.0 | 2.6 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 1.2 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 2.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 1.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 1.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.2 | GO:0021592 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.3 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 1.3 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.2 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 3.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.3 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.5 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 1.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.8 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 0.1 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.0 | 0.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.4 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 9.7 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.0 | 0.7 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.2 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.0 | 0.7 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.3 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.4 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.7 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.4 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.3 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.9 | GO:1901186 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 1.9 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.3 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.4 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 1.8 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.5 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.4 | GO:1902400 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 0.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.7 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 1.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.0 | GO:0007442 | hindgut morphogenesis(GO:0007442) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 13.7 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.7 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 2.3 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 6.8 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.5 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.0 | GO:0015883 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 2.1 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.3 | GO:0090662 | ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 1.1 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.0 | 0.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 1.2 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.8 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.1 | GO:0046368 | GDP-L-fucose biosynthetic process(GO:0042350) GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 2.3 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.3 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 1.2 | GO:0001892 | embryonic placenta development(GO:0001892) |
| 0.0 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.3 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.9 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 1.0 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.0 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.1 | GO:2001020 | regulation of response to DNA damage stimulus(GO:2001020) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 15.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 1.2 | 7.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.9 | 1.8 | GO:0044391 | ribosomal subunit(GO:0044391) |
| 0.9 | 5.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.7 | 5.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.7 | 6.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.6 | 3.9 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.6 | 3.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.5 | 4.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.5 | 6.9 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.4 | 2.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.4 | 8.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.4 | 6.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 5.9 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.4 | 4.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 4.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 10.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.3 | 1.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 1.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 2.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 4.5 | GO:0043219 | lateral loop(GO:0043219) |
| 0.3 | 0.8 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.2 | 2.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.2 | 1.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 0.7 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 0.7 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.2 | 2.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 2.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 2.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 2.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 0.9 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.2 | 0.9 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 1.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 2.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.2 | 1.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 2.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 1.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.2 | 1.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 3.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 1.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 1.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.6 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.2 | 1.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.2 | 1.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 8.1 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.9 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 2.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.6 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 1.6 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.7 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 2.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 4.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 2.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.8 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 0.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 3.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.0 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.8 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 4.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.7 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 2.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.5 | GO:0032279 | axonemal microtubule(GO:0005879) asymmetric synapse(GO:0032279) |
| 0.1 | 1.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 13.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 2.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.8 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.6 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 1.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 1.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 2.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 4.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 1.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.1 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.1 | 0.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 3.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 2.9 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 1.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 2.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 2.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.4 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 1.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 39.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 13.9 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 12.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 4.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 4.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 2.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.9 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 7.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.0 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 1.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 3.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 1.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 21.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.9 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.9 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 3.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 1.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 4.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.8 | GO:0030055 | cell-substrate adherens junction(GO:0005924) cell-substrate junction(GO:0030055) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 3.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 2.0 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.5 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 1.1 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 1.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 1.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.7 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 4.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 8.8 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 1.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 1.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.2 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 1.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.0 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 6.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.2 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 12.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.1 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.0 | 0.0 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 1.8 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 1.7 | 5.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 1.6 | 4.9 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 1.2 | 3.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.2 | 7.0 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 1.1 | 15.8 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.8 | 2.3 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.7 | 5.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.7 | 2.0 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.6 | 2.5 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.6 | 1.8 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.5 | 3.6 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.5 | 7.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.5 | 1.5 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.5 | 2.8 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.5 | 1.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.5 | 1.8 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.5 | 1.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.4 | 1.7 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.4 | 2.5 | GO:0070905 | serine binding(GO:0070905) |
| 0.4 | 1.6 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.4 | 2.0 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.4 | 1.9 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.4 | 1.8 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.4 | 1.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.4 | 1.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.4 | 3.6 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.4 | 1.8 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.4 | 1.8 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.3 | 1.0 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.3 | 1.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.3 | 1.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.3 | 8.2 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.3 | 1.0 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.3 | 1.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 2.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.3 | 1.5 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 10.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 0.9 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.3 | 1.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.3 | 1.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 2.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 1.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.3 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 1.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 1.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.3 | 3.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 4.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 3.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.0 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 4.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 1.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 1.6 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.2 | 0.7 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.2 | 1.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 2.0 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 2.0 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 1.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 9.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 0.8 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.2 | 3.6 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 6.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.5 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.2 | 1.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 1.0 | GO:0052813 | phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 1.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 1.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 3.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 0.7 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 2.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.7 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 1.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 1.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 3.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 1.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 0.3 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.2 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.2 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.2 | 4.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 1.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 2.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 0.5 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.2 | 0.5 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.2 | 0.5 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.2 | 0.5 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.1 | 3.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.7 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 1.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 3.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.8 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 1.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.3 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 1.6 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 4.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 2.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.1 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 3.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 1.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 11.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.3 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 2.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.6 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.6 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 6.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 1.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.5 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 4.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 0.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.6 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.9 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.7 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 3.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 3.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 2.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.4 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 1.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.6 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.5 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.4 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 2.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.7 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.7 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 1.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 3.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.4 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 1.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 4.0 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.6 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.0 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 3.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.9 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.2 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 1.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.6 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.3 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.7 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 2.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.3 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 1.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 5.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 2.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.3 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 7.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.9 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.7 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.1 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.1 | 0.2 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 0.6 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.6 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.3 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.1 | 1.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 1.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.7 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.7 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.3 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 1.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 4.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 3.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 1.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.5 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.7 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 2.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.4 | GO:0015925 | beta-galactosidase activity(GO:0004565) galactosidase activity(GO:0015925) |
| 0.0 | 1.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 1.0 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.9 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 2.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.7 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.8 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 2.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.4 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 5.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 2.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 4.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 14.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 5.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.0 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 4.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.3 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.9 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 5.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.0 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 2.7 | GO:0015103 | inorganic anion transmembrane transporter activity(GO:0015103) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.6 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 1.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 9.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.6 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 2.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 3.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 1.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.2 | 1.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 5.8 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 5.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 7.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 11.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 4.4 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 5.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 1.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 4.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 6.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 4.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 2.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 8.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 4.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 8.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 0.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 3.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 5.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 3.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.9 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 4.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 0.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 3.4 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 5.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 13.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 3.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 2.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.4 | 26.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 13.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.3 | 6.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 5.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 31.0 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.2 | 3.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 4.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 3.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 6.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 3.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 1.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 5.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 7.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 3.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 4.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 5.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 5.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 0.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.5 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 1.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 5.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 6.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 5.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 4.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.0 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 6.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.8 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 5.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 3.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.9 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 2.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 3.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME ORC1 REMOVAL FROM CHROMATIN | Genes involved in Orc1 removal from chromatin |
| 0.0 | 1.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 9.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME PURINE METABOLISM | Genes involved in Purine metabolism |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 3.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.7 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 1.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.0 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.7 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME INTERFERON SIGNALING | Genes involved in Interferon Signaling |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |