Project
Illumina Body Map 2
Navigation
Downloads
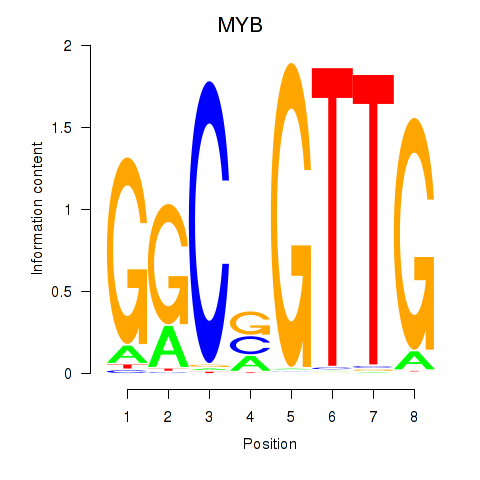
Results for MYB
Z-value: 3.66
Transcription factors associated with MYB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYB
|
ENSG00000118513.14 | MYB proto-oncogene, transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYB | hg19_v2_chr6_+_135502501_135502652 | 0.31 | 8.4e-02 | Click! |
Activity profile of MYB motif
Sorted Z-values of MYB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_54814288 | 13.10 |
ENST00000585477.1
|
BOD1L2
|
biorientation of chromosomes in cell division 1-like 2 |
| chr15_+_97326619 | 12.77 |
ENST00000328504.3
|
SPATA8
|
spermatogenesis associated 8 |
| chr9_-_104357277 | 12.16 |
ENST00000374806.1
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta |
| chr3_+_52444651 | 12.01 |
ENST00000327906.3
|
PHF7
|
PHD finger protein 7 |
| chr8_-_39695719 | 11.71 |
ENST00000347580.4
ENST00000379853.2 ENST00000521880.1 |
ADAM2
|
ADAM metallopeptidase domain 2 |
| chr6_+_22569784 | 11.48 |
ENST00000510882.2
|
HDGFL1
|
hepatoma derived growth factor-like 1 |
| chr22_-_20461786 | 10.86 |
ENST00000426804.1
|
RIMBP3
|
RIMS binding protein 3 |
| chr1_+_156308245 | 10.85 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr16_+_66613351 | 10.22 |
ENST00000379486.2
ENST00000268595.2 |
CMTM2
|
CKLF-like MARVEL transmembrane domain containing 2 |
| chr7_+_72742178 | 10.20 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr5_+_10441970 | 10.16 |
ENST00000274134.4
|
ROPN1L
|
rhophilin associated tail protein 1-like |
| chr4_+_155702496 | 10.13 |
ENST00000510397.1
|
RBM46
|
RNA binding motif protein 46 |
| chr10_+_32735030 | 10.06 |
ENST00000277657.6
ENST00000362006.5 |
CCDC7
|
coiled-coil domain containing 7 |
| chr15_+_97326659 | 9.97 |
ENST00000558553.1
|
SPATA8
|
spermatogenesis associated 8 |
| chr14_+_29236269 | 9.97 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr6_+_22569678 | 9.89 |
ENST00000230012.3
|
HDGFL1
|
hepatoma derived growth factor-like 1 |
| chr5_+_169659950 | 9.82 |
ENST00000593851.1
|
C5orf58
|
chromosome 5 open reading frame 58 |
| chr4_+_123300664 | 9.73 |
ENST00000388725.2
|
ADAD1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr4_+_44019074 | 9.67 |
ENST00000512678.1
|
RP11-328N19.1
|
RP11-328N19.1 |
| chr11_+_18433840 | 9.41 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr22_+_21737663 | 9.38 |
ENST00000434111.1
|
RIMBP3B
|
RIMS binding protein 3B |
| chr1_+_92414928 | 9.36 |
ENST00000362005.3
ENST00000370389.2 ENST00000399546.2 ENST00000423434.1 ENST00000394530.3 ENST00000440509.1 |
BRDT
|
bromodomain, testis-specific |
| chr8_-_39695764 | 9.06 |
ENST00000265708.4
|
ADAM2
|
ADAM metallopeptidase domain 2 |
| chr4_+_123300591 | 9.04 |
ENST00000439307.1
ENST00000388724.2 |
ADAD1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr4_+_123300121 | 8.81 |
ENST00000446706.1
ENST00000296513.2 |
ADAD1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr7_+_72742162 | 8.36 |
ENST00000431982.2
|
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr3_-_50383096 | 8.35 |
ENST00000442887.1
ENST00000360165.3 |
ZMYND10
|
zinc finger, MYND-type containing 10 |
| chr11_+_130184888 | 8.23 |
ENST00000602376.1
ENST00000532116.3 ENST00000602310.1 |
RP11-121M22.1
|
RP11-121M22.1 |
| chr2_+_132160448 | 8.09 |
ENST00000437751.1
|
AC073869.19
|
long intergenic non-protein coding RNA 1120 |
| chr1_+_156308403 | 7.85 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr12_-_54867352 | 7.67 |
ENST00000305879.5
|
GTSF1
|
gametocyte specific factor 1 |
| chr17_-_56565736 | 7.63 |
ENST00000323777.3
|
HSF5
|
heat shock transcription factor family member 5 |
| chr16_-_48189898 | 7.44 |
ENST00000527640.1
|
ABCC12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr1_+_53925063 | 7.43 |
ENST00000371445.3
|
DMRTB1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr11_-_123756334 | 7.31 |
ENST00000528595.1
ENST00000375026.2 |
TMEM225
|
transmembrane protein 225 |
| chr5_-_41071392 | 7.09 |
ENST00000399564.4
|
MROH2B
|
maestro heat-like repeat family member 2B |
| chr5_-_157079428 | 7.04 |
ENST00000265007.6
|
SOX30
|
SRY (sex determining region Y)-box 30 |
| chr17_-_33288419 | 7.03 |
ENST00000421975.3
|
CCT6B
|
chaperonin containing TCP1, subunit 6B (zeta 2) |
| chr17_+_58499844 | 6.96 |
ENST00000269127.4
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr5_-_157079372 | 6.95 |
ENST00000311371.5
|
SOX30
|
SRY (sex determining region Y)-box 30 |
| chr4_+_128651530 | 6.89 |
ENST00000281154.4
|
SLC25A31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chr17_-_33288522 | 6.86 |
ENST00000314144.5
|
CCT6B
|
chaperonin containing TCP1, subunit 6B (zeta 2) |
| chr17_-_7197881 | 6.85 |
ENST00000007699.5
|
YBX2
|
Y box binding protein 2 |
| chr3_-_172859017 | 6.81 |
ENST00000351008.3
|
SPATA16
|
spermatogenesis associated 16 |
| chr13_+_25338290 | 6.81 |
ENST00000255324.5
ENST00000381921.1 ENST00000255325.6 |
RNF17
|
ring finger protein 17 |
| chr17_-_33288467 | 6.67 |
ENST00000436961.3
|
CCT6B
|
chaperonin containing TCP1, subunit 6B (zeta 2) |
| chr19_+_35396219 | 6.58 |
ENST00000595783.1
|
CTC-523E23.6
|
CTC-523E23.6 |
| chr12_+_107078474 | 6.56 |
ENST00000552866.1
ENST00000229387.5 |
RFX4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr7_-_88425025 | 6.42 |
ENST00000297203.2
|
C7orf62
|
chromosome 7 open reading frame 62 |
| chr11_-_111383064 | 6.37 |
ENST00000525791.1
ENST00000456861.2 ENST00000356018.2 |
BTG4
|
B-cell translocation gene 4 |
| chr8_+_39442097 | 6.32 |
ENST00000265707.5
ENST00000379866.1 ENST00000520772.1 ENST00000541111.1 |
ADAM18
|
ADAM metallopeptidase domain 18 |
| chr17_-_3375033 | 6.27 |
ENST00000268981.5
ENST00000397168.3 ENST00000572969.1 ENST00000355380.4 ENST00000571553.1 ENST00000574797.1 ENST00000575375.1 |
SPATA22
|
spermatogenesis associated 22 |
| chr5_-_101834712 | 6.16 |
ENST00000506729.1
ENST00000389019.3 ENST00000379810.1 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr9_+_6328360 | 6.06 |
ENST00000381428.1
ENST00000314556.3 |
TPD52L3
|
tumor protein D52-like 3 |
| chr17_+_58499066 | 6.05 |
ENST00000474834.1
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr19_+_51754332 | 5.98 |
ENST00000601727.1
|
SIGLECL1
|
SIGLEC family like 1 |
| chr14_+_21785693 | 5.93 |
ENST00000382933.4
ENST00000557351.1 |
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr1_-_182922505 | 5.88 |
ENST00000367547.3
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like |
| chr11_-_68519026 | 5.79 |
ENST00000255087.5
|
MTL5
|
metallothionein-like 5, testis-specific (tesmin) |
| chr2_-_111435610 | 5.79 |
ENST00000447014.1
ENST00000420328.1 ENST00000535254.1 ENST00000409311.1 ENST00000302759.6 |
BUB1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr19_+_51754526 | 5.78 |
ENST00000596117.1
|
SIGLECL1
|
SIGLEC family like 1 |
| chr3_-_71834318 | 5.77 |
ENST00000353065.3
|
PROK2
|
prokineticin 2 |
| chr9_+_131219250 | 5.69 |
ENST00000470061.1
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr14_+_54863739 | 5.62 |
ENST00000541304.1
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr5_-_101834617 | 5.60 |
ENST00000513675.1
ENST00000379807.3 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr4_+_155702365 | 5.57 |
ENST00000514866.1
ENST00000281722.3 |
RBM46
|
RNA binding motif protein 46 |
| chr14_+_54863682 | 5.43 |
ENST00000543789.2
ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr7_+_100183927 | 5.41 |
ENST00000241071.6
ENST00000360609.2 |
FBXO24
|
F-box protein 24 |
| chr17_-_3417062 | 5.39 |
ENST00000570318.1
ENST00000541913.1 |
SPATA22
|
spermatogenesis associated 22 |
| chr14_+_54863667 | 5.31 |
ENST00000335183.6
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr22_-_21905120 | 5.17 |
ENST00000331505.5
|
RIMBP3C
|
RIMS binding protein 3C |
| chr22_+_30476163 | 5.14 |
ENST00000336726.6
|
HORMAD2
|
HORMA domain containing 2 |
| chr9_+_131219179 | 5.13 |
ENST00000372791.3
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr3_+_181670131 | 5.10 |
ENST00000482787.3
ENST00000600750.1 ENST00000596820.1 ENST00000490001.1 ENST00000467313.1 ENST00000474045.1 |
RP11-416O18.1
|
RP11-416O18.1 |
| chr13_-_52026730 | 5.04 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr15_+_69222827 | 5.03 |
ENST00000310673.3
ENST00000448182.3 ENST00000260364.5 |
SPESP1
NOX5
|
sperm equatorial segment protein 1 NADPH oxidase, EF-hand calcium binding domain 5 |
| chr19_-_35395961 | 5.02 |
ENST00000605047.1
|
CTC-523E23.15
|
CTC-523E23.15 |
| chr11_+_125774258 | 4.96 |
ENST00000263576.6
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr10_-_46167722 | 4.90 |
ENST00000374366.3
ENST00000344646.5 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr5_-_156593273 | 4.89 |
ENST00000302938.4
|
FAM71B
|
family with sequence similarity 71, member B |
| chr9_-_127269661 | 4.85 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr1_-_211848899 | 4.84 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chrX_+_102965835 | 4.80 |
ENST00000319560.6
|
TMEM31
|
transmembrane protein 31 |
| chr3_-_150421728 | 4.74 |
ENST00000295910.6
ENST00000491361.1 |
FAM194A
|
family with sequence similarity 194, member A |
| chr11_+_45743858 | 4.69 |
ENST00000532307.1
|
CTD-2210P24.1
|
CTD-2210P24.1 |
| chr17_-_39093672 | 4.65 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr15_+_74165945 | 4.58 |
ENST00000535547.2
ENST00000300504.2 ENST00000562056.1 |
TBC1D21
|
TBC1 domain family, member 21 |
| chr1_+_179561011 | 4.56 |
ENST00000294848.8
ENST00000444136.1 |
TDRD5
|
tudor domain containing 5 |
| chr12_+_29542557 | 4.55 |
ENST00000550906.1
|
OVCH1-AS1
|
OVCH1 antisense RNA 1 |
| chr15_+_73735490 | 4.52 |
ENST00000331090.6
ENST00000560581.1 |
C15orf60
|
chromosome 15 open reading frame 60 |
| chr19_+_36195429 | 4.51 |
ENST00000392197.2
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr9_+_6328338 | 4.47 |
ENST00000344545.5
|
TPD52L3
|
tumor protein D52-like 3 |
| chr13_-_103426112 | 4.46 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr3_-_59035673 | 4.44 |
ENST00000491845.1
ENST00000472469.1 ENST00000471288.1 ENST00000295966.7 |
C3orf67
|
chromosome 3 open reading frame 67 |
| chr8_-_101157680 | 4.42 |
ENST00000428847.2
|
FBXO43
|
F-box protein 43 |
| chr13_-_103426081 | 4.41 |
ENST00000376022.1
ENST00000376021.4 |
TEX30
|
testis expressed 30 |
| chr1_-_109656439 | 4.41 |
ENST00000369949.4
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr1_+_212208919 | 4.40 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr16_-_11363178 | 4.39 |
ENST00000312693.3
|
TNP2
|
transition protein 2 (during histone to protamine replacement) |
| chr19_+_33210645 | 4.37 |
ENST00000444215.2
|
TDRD12
|
tudor domain containing 12 |
| chr10_-_64028466 | 4.36 |
ENST00000395265.1
ENST00000373789.3 ENST00000395260.3 |
RTKN2
|
rhotekin 2 |
| chr17_-_8702667 | 4.32 |
ENST00000329805.4
|
MFSD6L
|
major facilitator superfamily domain containing 6-like |
| chr6_+_46655612 | 4.31 |
ENST00000544460.1
|
TDRD6
|
tudor domain containing 6 |
| chr11_+_45743931 | 4.30 |
ENST00000530051.1
|
CTD-2210P24.1
|
CTD-2210P24.1 |
| chr15_+_72690614 | 4.30 |
ENST00000341689.3
ENST00000567679.1 |
TMEM202
|
transmembrane protein 202 |
| chr4_-_57687847 | 4.30 |
ENST00000504762.1
ENST00000248701.4 ENST00000506738.1 |
SPINK2
|
serine peptidase inhibitor, Kazal type 2 (acrosin-trypsin inhibitor) |
| chr1_+_166958504 | 4.27 |
ENST00000447624.1
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chrY_+_22737678 | 4.27 |
ENST00000382772.3
|
EIF1AY
|
eukaryotic translation initiation factor 1A, Y-linked |
| chr14_+_104394770 | 4.27 |
ENST00000409874.4
ENST00000339063.5 |
TDRD9
|
tudor domain containing 9 |
| chr17_-_46894576 | 4.24 |
ENST00000393382.3
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6 |
| chr1_-_32827682 | 4.23 |
ENST00000432622.1
|
FAM229A
|
family with sequence similarity 229, member A |
| chr15_+_59499031 | 4.22 |
ENST00000307144.4
|
LDHAL6B
|
lactate dehydrogenase A-like 6B |
| chr11_-_116708302 | 4.13 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr12_-_6961050 | 4.12 |
ENST00000538862.2
|
CDCA3
|
cell division cycle associated 3 |
| chr17_-_76573465 | 4.08 |
ENST00000585328.1
ENST00000389840.5 |
DNAH17
|
dynein, axonemal, heavy chain 17 |
| chr6_+_167412835 | 4.05 |
ENST00000349556.4
|
FGFR1OP
|
FGFR1 oncogene partner |
| chr1_+_166958497 | 4.03 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr15_+_75491203 | 3.96 |
ENST00000562637.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr19_+_46236509 | 3.94 |
ENST00000457052.2
|
AC074212.3
|
Uncharacterized protein |
| chr17_-_60885645 | 3.92 |
ENST00000544856.2
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr17_-_60885659 | 3.88 |
ENST00000311269.5
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr19_-_16738984 | 3.87 |
ENST00000600060.1
ENST00000263390.3 |
MED26
|
mediator complex subunit 26 |
| chr3_-_52188397 | 3.87 |
ENST00000474012.1
ENST00000296484.2 |
POC1A
|
POC1 centriolar protein A |
| chr17_-_60885700 | 3.86 |
ENST00000583600.1
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chrX_-_70128580 | 3.81 |
ENST00000374333.2
|
TEX11
|
testis expressed 11 |
| chr1_-_85155939 | 3.78 |
ENST00000603677.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr20_-_58508702 | 3.76 |
ENST00000357552.3
ENST00000425931.1 |
SYCP2
|
synaptonemal complex protein 2 |
| chr7_-_72992663 | 3.72 |
ENST00000432538.1
|
TBL2
|
transducin (beta)-like 2 |
| chr6_-_27279949 | 3.71 |
ENST00000444565.1
ENST00000377451.2 |
POM121L2
|
POM121 transmembrane nucleoporin-like 2 |
| chr13_+_25670268 | 3.71 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr22_+_51176624 | 3.70 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr17_+_18647326 | 3.67 |
ENST00000395667.1
ENST00000395665.4 ENST00000308799.4 ENST00000301938.4 |
FBXW10
|
F-box and WD repeat domain containing 10 |
| chrX_-_125686784 | 3.63 |
ENST00000371126.1
|
DCAF12L1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr1_+_43824669 | 3.62 |
ENST00000372462.1
|
CDC20
|
cell division cycle 20 |
| chr1_-_112106578 | 3.61 |
ENST00000369717.4
|
ADORA3
|
adenosine A3 receptor |
| chr1_+_43824577 | 3.60 |
ENST00000310955.6
|
CDC20
|
cell division cycle 20 |
| chr1_-_109203997 | 3.56 |
ENST00000370032.5
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr3_-_71834207 | 3.55 |
ENST00000295619.3
|
PROK2
|
prokineticin 2 |
| chr22_-_22090043 | 3.54 |
ENST00000403503.1
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chr1_+_26517052 | 3.54 |
ENST00000338855.2
ENST00000456354.2 |
CATSPER4
|
cation channel, sperm associated 4 |
| chr1_+_41707996 | 3.49 |
ENST00000425554.1
|
RP11-399E6.1
|
RP11-399E6.1 |
| chr15_+_40886439 | 3.48 |
ENST00000532056.1
ENST00000399668.2 |
CASC5
|
cancer susceptibility candidate 5 |
| chr7_-_123174610 | 3.47 |
ENST00000324698.6
ENST00000434450.1 |
IQUB
|
IQ motif and ubiquitin domain containing |
| chr1_+_244624678 | 3.47 |
ENST00000366534.4
ENST00000366533.4 ENST00000428042.1 ENST00000366531.3 |
C1orf101
|
chromosome 1 open reading frame 101 |
| chr15_-_71407806 | 3.47 |
ENST00000566432.1
ENST00000567117.1 |
CT62
|
cancer/testis antigen 62 |
| chr11_+_85566422 | 3.42 |
ENST00000342404.3
|
CCDC83
|
coiled-coil domain containing 83 |
| chr1_-_112106556 | 3.41 |
ENST00000443498.1
|
ADORA3
|
adenosine A3 receptor |
| chr15_-_71407833 | 3.40 |
ENST00000449977.2
|
CT62
|
cancer/testis antigen 62 |
| chr14_+_94640633 | 3.39 |
ENST00000304338.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr11_+_125774362 | 3.38 |
ENST00000530414.1
ENST00000530129.2 |
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr1_+_92414952 | 3.38 |
ENST00000449584.1
ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT
|
bromodomain, testis-specific |
| chr4_-_186392843 | 3.37 |
ENST00000393540.3
ENST00000506876.1 |
CCDC110
|
coiled-coil domain containing 110 |
| chr15_+_84116106 | 3.34 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr15_+_93124059 | 3.33 |
ENST00000555864.1
|
RP11-386M24.3
|
RP11-386M24.3 |
| chr14_+_94385235 | 3.33 |
ENST00000557719.1
ENST00000267594.5 |
FAM181A
|
family with sequence similarity 181, member A |
| chr20_-_34117447 | 3.27 |
ENST00000246199.2
ENST00000424444.1 ENST00000374345.4 ENST00000444723.1 |
C20orf173
|
chromosome 20 open reading frame 173 |
| chr11_+_6255995 | 3.27 |
ENST00000533426.1
|
CNGA4
|
cyclic nucleotide gated channel alpha 4 |
| chr7_-_72992865 | 3.27 |
ENST00000452475.1
|
TBL2
|
transducin (beta)-like 2 |
| chrX_-_70128554 | 3.27 |
ENST00000395889.2
|
TEX11
|
testis expressed 11 |
| chr22_-_46659219 | 3.26 |
ENST00000253255.5
|
PKDREJ
|
polycystin (PKD) family receptor for egg jelly |
| chr19_-_9609272 | 3.26 |
ENST00000301480.4
|
ZNF560
|
zinc finger protein 560 |
| chr1_-_235098935 | 3.23 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr7_+_48075108 | 3.22 |
ENST00000420324.1
ENST00000435376.1 ENST00000430738.1 ENST00000348904.3 ENST00000539619.1 |
C7orf57
|
chromosome 7 open reading frame 57 |
| chrX_-_102531717 | 3.22 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chrX_-_151922340 | 3.21 |
ENST00000370284.1
ENST00000543232.1 ENST00000393876.1 ENST00000393872.3 |
MAGEA2
|
melanoma antigen family A, 2 |
| chr2_+_26624775 | 3.21 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr3_+_113666748 | 3.19 |
ENST00000330212.3
ENST00000498275.1 |
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr17_+_8243154 | 3.18 |
ENST00000328248.2
ENST00000584943.1 |
ODF4
|
outer dense fiber of sperm tails 4 |
| chr13_-_60737898 | 3.16 |
ENST00000377908.2
ENST00000400319.1 ENST00000400320.1 ENST00000267215.4 |
DIAPH3
|
diaphanous-related formin 3 |
| chr9_-_23821273 | 3.16 |
ENST00000380110.4
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr17_-_58499698 | 3.14 |
ENST00000590297.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr7_-_72993033 | 3.14 |
ENST00000305632.5
|
TBL2
|
transducin (beta)-like 2 |
| chr17_-_46262541 | 3.12 |
ENST00000579336.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr15_+_45722727 | 3.11 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr7_+_102105370 | 3.10 |
ENST00000292616.5
|
LRWD1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr11_+_93063878 | 3.09 |
ENST00000298050.3
|
CCDC67
|
coiled-coil domain containing 67 |
| chr6_+_44126545 | 3.09 |
ENST00000532171.1
ENST00000398776.1 ENST00000542245.1 |
CAPN11
|
calpain 11 |
| chr1_-_68962782 | 3.06 |
ENST00000456315.2
|
DEPDC1
|
DEP domain containing 1 |
| chr16_+_66600294 | 3.06 |
ENST00000535705.1
ENST00000332695.7 ENST00000336328.6 ENST00000528324.1 ENST00000531885.1 ENST00000529506.1 ENST00000457188.2 ENST00000533666.1 ENST00000533953.1 ENST00000379500.2 ENST00000328020.6 |
CMTM1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr3_-_128712906 | 3.05 |
ENST00000511438.1
|
KIAA1257
|
KIAA1257 |
| chrX_+_151883090 | 3.04 |
ENST00000370293.2
ENST00000423993.1 ENST00000447530.1 ENST00000458057.1 ENST00000331220.2 ENST00000422085.1 ENST00000453150.1 ENST00000409560.1 |
MAGEA2B
|
melanoma antigen family A, 2B |
| chr19_+_38826415 | 3.04 |
ENST00000410018.1
ENST00000409235.3 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chr19_+_827823 | 3.04 |
ENST00000233997.2
|
AZU1
|
azurocidin 1 |
| chr7_+_53103320 | 3.04 |
ENST00000408890.4
|
POM121L12
|
POM121 transmembrane nucleoporin-like 12 |
| chr18_-_6929797 | 3.03 |
ENST00000581725.1
ENST00000583316.1 |
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr4_-_141348789 | 2.99 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chrX_+_136648297 | 2.99 |
ENST00000287538.5
|
ZIC3
|
Zic family member 3 |
| chr5_-_127674883 | 2.98 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr12_+_49297899 | 2.97 |
ENST00000552942.1
ENST00000320516.4 |
CCDC65
|
coiled-coil domain containing 65 |
| chr17_+_42733803 | 2.94 |
ENST00000409122.2
|
C17orf104
|
chromosome 17 open reading frame 104 |
| chr17_+_40950900 | 2.94 |
ENST00000588527.1
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr2_+_149974684 | 2.94 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr7_+_62809239 | 2.94 |
ENST00000456890.1
|
AC006455.1
|
AC006455.1 |
| chr1_-_85156090 | 2.93 |
ENST00000605755.1
ENST00000437941.2 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr19_+_1104415 | 2.93 |
ENST00000585362.2
|
GPX4
|
glutathione peroxidase 4 |
| chr7_-_72742085 | 2.92 |
ENST00000333149.2
|
TRIM50
|
tripartite motif containing 50 |
| chr19_+_36195467 | 2.90 |
ENST00000426659.2
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr1_-_68962744 | 2.88 |
ENST00000525124.1
|
DEPDC1
|
DEP domain containing 1 |
| chr11_-_68518910 | 2.87 |
ENST00000544963.1
ENST00000443940.2 |
MTL5
|
metallothionein-like 5, testis-specific (tesmin) |
| chr9_-_127269488 | 2.87 |
ENST00000455734.1
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.7 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 3.1 | 9.4 | GO:0019516 | lactate oxidation(GO:0019516) |
| 2.9 | 11.7 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 1.7 | 6.9 | GO:0009386 | translational attenuation(GO:0009386) |
| 1.7 | 5.0 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 1.6 | 42.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 1.5 | 7.7 | GO:0007538 | primary sex determination(GO:0007538) |
| 1.4 | 5.8 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.3 | 1.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 1.2 | 6.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 1.2 | 7.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 1.1 | 9.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 1.1 | 10.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 1.1 | 20.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 1.0 | 4.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 1.0 | 3.0 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 1.0 | 8.0 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 1.0 | 4.8 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.9 | 2.7 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.9 | 5.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.8 | 3.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.8 | 2.3 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.7 | 5.2 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.7 | 7.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.7 | 3.6 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.7 | 8.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.7 | 7.6 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.7 | 2.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.6 | 2.5 | GO:0072301 | negative regulation of metanephric glomerulus development(GO:0072299) regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.6 | 19.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.6 | 1.9 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.6 | 1.8 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.6 | 4.2 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.6 | 7.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.6 | 6.6 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.6 | 1.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.5 | 10.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.5 | 5.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.5 | 2.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.5 | 2.0 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.5 | 1.5 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.5 | 0.5 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.5 | 2.4 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.4 | 2.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.4 | 2.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.4 | 8.2 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.4 | 2.4 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.4 | 3.2 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.4 | 4.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 1.2 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.4 | 9.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.4 | 1.6 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.4 | 77.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.4 | 9.7 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.4 | 2.5 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.4 | 1.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.4 | 4.3 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.4 | 1.8 | GO:0014735 | regulation of muscle atrophy(GO:0014735) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.3 | 8.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.3 | 3.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 1.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.3 | 6.5 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.3 | 6.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.3 | 1.3 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.3 | 1.0 | GO:1900159 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.3 | 2.6 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.3 | 2.5 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.3 | 1.8 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.3 | 0.6 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.3 | 9.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.3 | 1.5 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.3 | 2.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 4.4 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.3 | 7.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.3 | 0.9 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.3 | 1.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.3 | 4.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.3 | 0.9 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.3 | 11.0 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 | 5.3 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.3 | 1.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.3 | 3.0 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 1.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.3 | 1.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.3 | 0.8 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.3 | 4.4 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.3 | 1.0 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.3 | 2.6 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.3 | 2.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.0 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.2 | 2.9 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 1.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.2 | 4.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 0.7 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.2 | 1.1 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.2 | 0.9 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.2 | 1.5 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.2 | 0.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 10.7 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.2 | 1.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 11.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 3.8 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 0.8 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.2 | 0.6 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.2 | 0.6 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.2 | 8.8 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.2 | 1.8 | GO:0003341 | cilium movement(GO:0003341) |
| 0.2 | 0.8 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 | 1.0 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.2 | 2.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 0.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.2 | 0.7 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 0.9 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 0.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 2.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 0.9 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.2 | 0.2 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.2 | 0.3 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.2 | 3.0 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.2 | 0.8 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.2 | 0.8 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 | 7.1 | GO:0007099 | centriole replication(GO:0007099) |
| 0.2 | 1.0 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 4.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 1.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.2 | 0.5 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 1.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 0.8 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.2 | 1.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 3.4 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 1.5 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.7 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 2.6 | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.1 | 1.9 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.3 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 2.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 2.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.5 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.9 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.5 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 2.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.8 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.6 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 4.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.9 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.4 | GO:1901859 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.1 | 3.9 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 2.1 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.5 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.1 | 0.6 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.5 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 7.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 4.1 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.5 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.9 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.3 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.1 | 3.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 3.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 1.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 2.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 6.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.4 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.1 | 1.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 2.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 1.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 12.5 | GO:1904029 | regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.5 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 2.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 2.1 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.1 | 1.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 2.7 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 2.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.5 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.1 | 0.3 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 1.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 1.9 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.1 | 1.9 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 2.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 42.7 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 2.1 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 1.6 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 3.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.4 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.4 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.3 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 1.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 0.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.1 | 0.5 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 1.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.5 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 1.2 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 5.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 3.0 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 1.0 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.7 | GO:1900116 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.4 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 1.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.1 | GO:2000349 | negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.2 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.2 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.5 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 1.0 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.8 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 1.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.7 | GO:0046851 | negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.0 | 2.9 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 1.6 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 1.7 | GO:0048265 | response to pain(GO:0048265) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 2.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 5.3 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.3 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.6 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 1.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.6 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.9 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.7 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 1.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.0 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.3 | GO:0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) |
| 0.0 | 0.2 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.0 | 0.6 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 1.9 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 1.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.3 | GO:0060444 | branching involved in mammary gland duct morphogenesis(GO:0060444) |
| 0.0 | 2.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 2.6 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.8 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.3 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.3 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:2001020 | regulation of response to DNA damage stimulus(GO:2001020) |
| 0.0 | 0.0 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.3 | GO:0030849 | autosome(GO:0030849) |
| 1.5 | 11.7 | GO:0005694 | chromosome(GO:0005694) |
| 1.4 | 4.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.2 | 3.7 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.1 | 9.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.1 | 11.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 1.1 | 4.4 | GO:1990923 | PET complex(GO:1990923) |
| 1.1 | 17.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.0 | 5.8 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.9 | 14.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.8 | 8.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.8 | 8.5 | GO:0000801 | central element(GO:0000801) |
| 0.8 | 3.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.7 | 4.3 | GO:0071547 | piP-body(GO:0071547) |
| 0.7 | 17.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.7 | 2.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.7 | 2.6 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.6 | 28.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.6 | 1.9 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.6 | 8.1 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.6 | 1.8 | GO:0001534 | radial spoke(GO:0001534) |
| 0.6 | 9.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.5 | 3.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 1.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.4 | 1.8 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 4.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.4 | 4.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.4 | 4.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.4 | 2.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 2.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.4 | 4.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 14.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 8.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.3 | 7.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.3 | 2.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 2.4 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 3.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.3 | 4.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.3 | 3.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 4.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 0.8 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.2 | 8.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 5.0 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.2 | 9.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 0.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 0.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.2 | 2.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 0.9 | GO:0032449 | CBM complex(GO:0032449) |
| 0.2 | 0.8 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.2 | 0.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.2 | 22.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.2 | 2.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 6.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 1.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 0.5 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.2 | 21.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 2.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 6.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 17.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 6.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.0 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 4.3 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 1.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 12.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 3.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 10.3 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 1.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 3.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.5 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 3.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 4.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 2.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 2.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 4.5 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 2.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 7.6 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 11.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 12.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 4.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 2.2 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 16.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 2.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 3.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 1.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 2.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.4 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0030286 | dynein complex(GO:0030286) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 27.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 1.0 | 4.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.0 | 10.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.0 | 5.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 1.0 | 4.0 | GO:0004040 | amidase activity(GO:0004040) |
| 1.0 | 2.9 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.9 | 7.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.7 | 6.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.7 | 6.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.6 | 4.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.6 | 3.0 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.6 | 3.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.5 | 18.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 2.7 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.5 | 2.5 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.5 | 25.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.5 | 2.0 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.5 | 2.0 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.5 | 1.5 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.5 | 3.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.5 | 1.8 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.4 | 7.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.4 | 2.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.4 | 1.2 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.4 | 5.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.4 | 14.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.3 | 1.4 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.3 | 3.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 1.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.3 | 2.7 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 3.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 0.9 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.3 | 1.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.3 | 0.8 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.2 | 4.6 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 11.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 2.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 11.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 0.9 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 7.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 1.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 0.6 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.2 | 11.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.2 | 10.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 2.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 1.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 0.7 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 6.6 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.2 | 2.9 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 2.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 13.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 0.7 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.2 | 2.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 5.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 0.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 0.8 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.2 | 10.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.8 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.7 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 2.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 4.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 23.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 2.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 2.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 5.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 1.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 11.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 8.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 3.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 7.9 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 9.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.9 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 3.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 4.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 2.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 6.0 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.3 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 8.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 2.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 2.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 3.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.9 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 1.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 2.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 1.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 3.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 2.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 3.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 10.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.5 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 5.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 23.1 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 30.5 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 1.0 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 3.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 2.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 3.4 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 2.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 11.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 2.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 13.1 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 33.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 19.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 16.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 7.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 5.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 11.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 3.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 5.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 3.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 2.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 7.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 5.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 10.6 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.2 | 25.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 5.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 25.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 7.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 2.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 4.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 4.4 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 2.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 4.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 2.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 5.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.8 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 2.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.6 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.2 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 9.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.3 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 2.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |