Project
Illumina Body Map 2
Navigation
Downloads
Results for MYBL2
Z-value: 2.74
Transcription factors associated with MYBL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYBL2
|
ENSG00000101057.11 | MYB proto-oncogene like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYBL2 | hg19_v2_chr20_+_42295745_42295797 | 0.59 | 3.6e-04 | Click! |
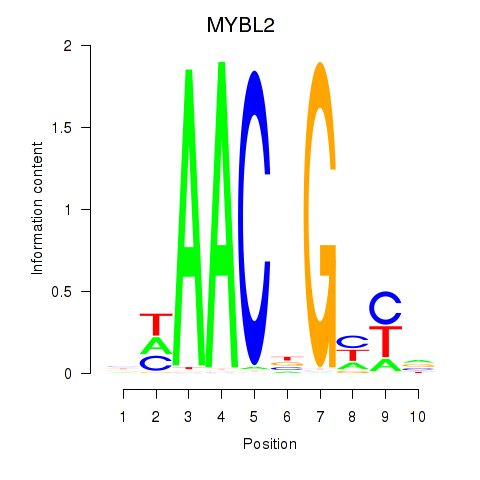
Activity profile of MYBL2 motif
Sorted Z-values of MYBL2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYBL2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 3.8 | 11.5 | GO:0019516 | lactate oxidation(GO:0019516) |
| 3.3 | 13.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 3.1 | 9.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 1.3 | 35.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 1.2 | 6.2 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 1.2 | 3.6 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 1.2 | 3.5 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 1.1 | 3.2 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 1.0 | 3.0 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 1.0 | 2.9 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.9 | 11.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 1.7 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.8 | 3.4 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.8 | 2.3 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.7 | 2.2 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.7 | 5.9 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.7 | 12.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.7 | 6.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.7 | 2.7 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.7 | 12.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.7 | 4.6 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.7 | 3.3 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.6 | 6.8 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.5 | 2.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.5 | 1.6 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.5 | 2.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.5 | 1.6 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.5 | 5.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.5 | 3.8 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.5 | 2.3 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.5 | 9.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.5 | 3.6 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.4 | 7.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.4 | 3.5 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.4 | 19.7 | GO:0007099 | centriole replication(GO:0007099) |
| 0.4 | 5.9 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.4 | 1.7 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.4 | 1.7 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.4 | 12.7 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.4 | 4.1 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.4 | 1.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.4 | 1.8 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 | 1.0 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.3 | 4.7 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.3 | 1.3 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.3 | 1.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.3 | 17.9 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.3 | 2.7 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.3 | 0.9 | GO:1904781 | regulation of protein localization to centrosome(GO:1904779) positive regulation of protein localization to centrosome(GO:1904781) |
| 0.3 | 1.7 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.3 | 3.8 | GO:0042148 | strand invasion(GO:0042148) |
| 0.3 | 5.5 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.3 | 0.8 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.2 | 2.5 | GO:0003352 | regulation of cilium movement(GO:0003352) microtubule severing(GO:0051013) |
| 0.2 | 1.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 1.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 1.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.2 | 1.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 9.0 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 4.9 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 3.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.2 | 2.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 1.9 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.2 | 0.8 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.2 | 0.7 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 1.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 2.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.2 | 4.4 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.2 | 2.9 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 0.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 24.3 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.2 | 3.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 28.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 0.6 | GO:0098942 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.2 | 0.8 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.2 | 4.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.4 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.9 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 | 3.0 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.9 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 1.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.1 | 6.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 2.7 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 4.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.5 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 4.1 | GO:0045143 | homologous chromosome segregation(GO:0045143) |
| 0.1 | 2.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 2.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 2.7 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.1 | 0.8 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 0.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 1.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 2.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 2.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.7 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 3.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 1.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 2.8 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.5 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 2.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.9 | GO:0009304 | transcription initiation from RNA polymerase III promoter(GO:0006384) tRNA transcription(GO:0009304) |
| 0.1 | 0.2 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.1 | 3.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 4.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.1 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.3 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.5 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.2 | GO:2000409 | astrocyte chemotaxis(GO:0035700) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) positive regulation of T cell extravasation(GO:2000409) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.1 | 3.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.3 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 1.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 1.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 5.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 1.8 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.1 | 3.7 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 2.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.8 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 1.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.2 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 1.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.7 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.5 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 3.6 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.4 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 1.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.4 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 3.9 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.6 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.8 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.5 | GO:0042723 | thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.4 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 2.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 1.1 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.1 | GO:0060537 | striated muscle tissue development(GO:0014706) muscle tissue development(GO:0060537) |
| 0.0 | 1.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 1.0 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 3.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.3 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.1 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 1.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 1.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0006473 | protein acetylation(GO:0006473) |
| 0.0 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 2.1 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.6 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.3 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.9 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.9 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 1.1 | GO:0006195 | purine nucleotide catabolic process(GO:0006195) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:0030849 | autosome(GO:0030849) |
| 1.3 | 4.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.3 | 3.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.0 | 8.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.9 | 8.8 | GO:0000801 | central element(GO:0000801) |
| 0.9 | 8.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.7 | 2.9 | GO:1990923 | PET complex(GO:1990923) |
| 0.7 | 4.3 | GO:0071547 | piP-body(GO:0071547) |
| 0.7 | 2.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.7 | 2.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.7 | 6.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.6 | 4.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.6 | 3.0 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.6 | 9.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.6 | 8.7 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.5 | 6.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.5 | 17.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.4 | 1.8 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 4.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.4 | 4.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.4 | 0.4 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.3 | 1.0 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.3 | 13.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 14.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.3 | 2.9 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.3 | 9.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 1.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.3 | 2.8 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 4.0 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.2 | 1.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 0.9 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 0.9 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.2 | 9.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 3.9 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 12.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 1.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.6 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 3.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.8 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 2.6 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 3.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 7.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.8 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 24.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 16.3 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 4.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 1.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.5 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 1.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 8.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 3.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 2.8 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.2 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 1.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.7 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.5 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 2.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 2.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 3.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.9 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.0 | 1.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 3.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 2.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 3.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 16.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 4.3 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 1.1 | 11.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.1 | 12.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 1.1 | 3.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 1.0 | 3.8 | GO:0004040 | amidase activity(GO:0004040) |
| 0.9 | 3.7 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.7 | 13.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.5 | 2.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.4 | 3.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.4 | 2.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.4 | 9.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 3.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.4 | 1.8 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.4 | 18.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.4 | 3.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.3 | 1.7 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.3 | 1.7 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.3 | 1.7 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.3 | 2.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.9 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.3 | 10.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.3 | 6.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 2.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.3 | 0.8 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.3 | 2.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 0.7 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 4.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 9.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.2 | 1.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 1.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 0.9 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.2 | 11.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 0.9 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 6.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 2.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 1.0 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.2 | 3.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 1.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.2 | 7.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 11.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 13.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 2.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 2.3 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.2 | 0.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 3.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 3.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 4.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.8 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 3.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 2.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.0 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 7.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 4.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 2.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.3 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.1 | 13.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.7 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 2.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 2.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 11.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.4 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 3.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 1.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 2.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 2.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.0 | 2.9 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 2.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 2.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 6.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 3.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 9.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 3.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 21.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 14.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 4.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 6.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 0.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 4.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 3.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 5.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 16.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 7.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 3.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 6.2 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 2.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 2.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 14.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 13.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 4.2 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.5 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 1.8 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.1 | 2.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 4.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 3.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |