Project
Illumina Body Map 2
Navigation
Downloads
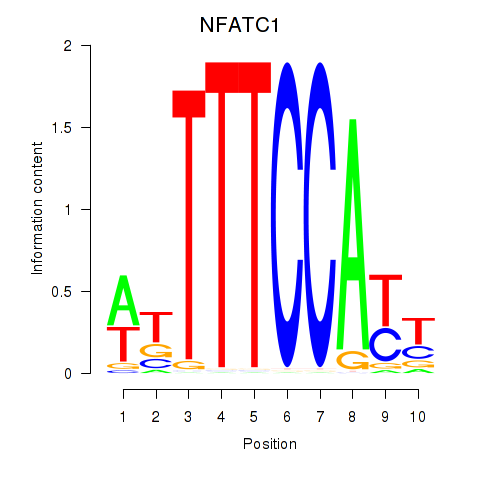
Results for NFATC1
Z-value: 1.72
Transcription factors associated with NFATC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC1
|
ENSG00000131196.13 | nuclear factor of activated T cells 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC1 | hg19_v2_chr18_+_77155856_77155939 | 0.55 | 1.0e-03 | Click! |
Activity profile of NFATC1 motif
Sorted Z-values of NFATC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_71532339 | 7.22 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr4_-_71532601 | 7.20 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr4_-_71532668 | 6.70 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_-_153517473 | 6.23 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr4_-_71532207 | 6.14 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr8_-_107782463 | 5.23 |
ENST00000311955.3
|
ABRA
|
actin-binding Rho activating protein |
| chr7_+_79998864 | 4.48 |
ENST00000435819.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr14_-_107013465 | 3.79 |
ENST00000390625.2
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr2_+_1417228 | 3.42 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr6_+_41021027 | 3.38 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr12_-_92536433 | 3.09 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr17_-_39781054 | 3.07 |
ENST00000463128.1
|
KRT17
|
keratin 17 |
| chr17_-_39780634 | 3.05 |
ENST00000577817.2
|
KRT17
|
keratin 17 |
| chr1_-_171621815 | 3.04 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr4_-_74847800 | 3.04 |
ENST00000296029.3
|
PF4
|
platelet factor 4 |
| chr7_+_15728003 | 3.01 |
ENST00000442176.1
|
AC005550.4
|
AC005550.4 |
| chr9_-_37034028 | 2.87 |
ENST00000520281.1
ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5
|
paired box 5 |
| chr9_+_124062071 | 2.84 |
ENST00000373818.4
|
GSN
|
gelsolin |
| chr12_-_114843889 | 2.81 |
ENST00000405440.2
|
TBX5
|
T-box 5 |
| chr2_+_204732666 | 2.70 |
ENST00000295854.6
ENST00000472206.1 |
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr5_+_118691008 | 2.60 |
ENST00000504642.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr10_-_92681033 | 2.58 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr8_-_124553437 | 2.57 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr1_+_87595497 | 2.56 |
ENST00000471417.1
|
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr17_-_39743139 | 2.52 |
ENST00000167586.6
|
KRT14
|
keratin 14 |
| chr17_-_46657473 | 2.48 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr3_+_157261116 | 2.39 |
ENST00000468043.1
ENST00000459838.1 ENST00000461040.1 ENST00000449199.2 ENST00000426338.2 |
C3orf55
|
chromosome 3 open reading frame 55 |
| chr1_+_87595433 | 2.37 |
ENST00000469312.2
ENST00000490006.2 |
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr7_+_28452130 | 2.36 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr1_-_186649543 | 2.34 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_-_80548493 | 2.32 |
ENST00000536800.1
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr5_+_118690466 | 2.32 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr1_-_203320617 | 2.31 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chr3_-_111852061 | 2.26 |
ENST00000488580.1
ENST00000460387.2 ENST00000484193.1 ENST00000487901.1 |
GCSAM
|
germinal center-associated, signaling and motility |
| chr1_-_226926864 | 2.21 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr5_-_158526693 | 2.15 |
ENST00000380654.4
|
EBF1
|
early B-cell factor 1 |
| chr2_-_211179883 | 2.15 |
ENST00000352451.3
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr12_-_52887034 | 2.12 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr2_-_161349909 | 2.11 |
ENST00000392753.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr2_+_204732487 | 2.09 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr14_-_92247032 | 2.08 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr5_-_158526756 | 2.08 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr2_+_75061108 | 2.06 |
ENST00000290573.2
|
HK2
|
hexokinase 2 |
| chr2_+_36923933 | 2.03 |
ENST00000497382.1
ENST00000404084.1 ENST00000379241.3 ENST00000401530.1 |
VIT
|
vitrin |
| chr3_+_130279178 | 2.01 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr10_-_121296045 | 2.01 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr7_-_83824169 | 1.97 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr6_+_32407619 | 1.97 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr3_-_111852128 | 1.96 |
ENST00000308910.4
|
GCSAM
|
germinal center-associated, signaling and motility |
| chrX_+_15518923 | 1.95 |
ENST00000348343.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr17_-_39677971 | 1.93 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr22_-_19512893 | 1.92 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr14_-_24977457 | 1.90 |
ENST00000250378.3
ENST00000206446.4 |
CMA1
|
chymase 1, mast cell |
| chr17_-_39780819 | 1.90 |
ENST00000311208.8
|
KRT17
|
keratin 17 |
| chr4_+_110834033 | 1.88 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr6_-_32920794 | 1.88 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr7_-_83824449 | 1.87 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_+_86046433 | 1.86 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr3_-_100565249 | 1.85 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr2_-_211168332 | 1.85 |
ENST00000341685.4
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr6_+_106534192 | 1.84 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr5_+_32788945 | 1.82 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr13_-_49975632 | 1.80 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chrX_+_133941218 | 1.79 |
ENST00000370784.4
ENST00000370785.3 |
FAM122C
|
family with sequence similarity 122C |
| chr12_+_52430894 | 1.75 |
ENST00000546842.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr4_+_169633310 | 1.73 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_-_218843623 | 1.72 |
ENST00000413280.1
|
TNS1
|
tensin 1 |
| chr3_-_111314230 | 1.71 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr8_-_13372253 | 1.70 |
ENST00000316609.5
|
DLC1
|
deleted in liver cancer 1 |
| chr3_+_14444063 | 1.68 |
ENST00000454876.2
ENST00000360861.3 ENST00000416216.2 |
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr1_+_209941942 | 1.67 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr18_-_53177984 | 1.67 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr2_+_36923901 | 1.67 |
ENST00000457137.2
|
VIT
|
vitrin |
| chr18_-_53257027 | 1.66 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr14_-_92413353 | 1.63 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr9_+_103340354 | 1.60 |
ENST00000307584.5
|
MURC
|
muscle-related coiled-coil protein |
| chr1_+_115642293 | 1.58 |
ENST00000448680.1
|
RP4-666F24.3
|
RP4-666F24.3 |
| chr2_-_40679148 | 1.57 |
ENST00000417271.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr22_+_38071615 | 1.57 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr2_-_163099546 | 1.57 |
ENST00000447386.1
|
FAP
|
fibroblast activation protein, alpha |
| chr4_-_87770416 | 1.55 |
ENST00000273905.6
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
| chr1_-_205904950 | 1.54 |
ENST00000340781.4
|
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr5_-_94417562 | 1.51 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr9_-_20621834 | 1.51 |
ENST00000429426.2
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr11_+_131240373 | 1.46 |
ENST00000374791.3
ENST00000436745.1 |
NTM
|
neurotrimin |
| chr3_+_152017360 | 1.45 |
ENST00000485910.1
ENST00000463374.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr18_+_77155856 | 1.45 |
ENST00000253506.5
ENST00000591814.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr15_-_90222642 | 1.44 |
ENST00000430628.2
|
PLIN1
|
perilipin 1 |
| chr1_-_169599314 | 1.44 |
ENST00000367786.2
ENST00000458599.2 ENST00000367795.2 ENST00000263686.6 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr15_-_90222610 | 1.44 |
ENST00000300055.5
|
PLIN1
|
perilipin 1 |
| chr3_-_100551141 | 1.43 |
ENST00000478235.1
ENST00000471901.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr14_-_69261310 | 1.43 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr1_+_162602244 | 1.43 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr4_-_90229142 | 1.42 |
ENST00000609438.1
|
GPRIN3
|
GPRIN family member 3 |
| chr10_+_11207088 | 1.41 |
ENST00000608830.1
|
CELF2
|
CUGBP, Elav-like family member 2 |
| chrX_-_19905703 | 1.40 |
ENST00000397821.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr1_-_169599353 | 1.39 |
ENST00000367793.2
ENST00000367794.2 ENST00000367792.2 ENST00000367791.2 ENST00000367788.2 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr1_-_150780757 | 1.38 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr1_+_78354243 | 1.38 |
ENST00000294624.8
|
NEXN
|
nexilin (F actin binding protein) |
| chrX_-_15332665 | 1.35 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr14_+_105047478 | 1.35 |
ENST00000410013.1
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr12_+_26348582 | 1.34 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr3_-_158390282 | 1.31 |
ENST00000264265.3
|
LXN
|
latexin |
| chr1_+_87595542 | 1.30 |
ENST00000461990.1
|
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr13_-_99667960 | 1.29 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr3_+_46395219 | 1.29 |
ENST00000445132.2
ENST00000292301.4 |
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr11_-_111794446 | 1.28 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr3_+_152017924 | 1.27 |
ENST00000465907.2
ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr13_+_73629107 | 1.26 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr21_-_27945562 | 1.24 |
ENST00000299340.4
ENST00000435845.2 |
CYYR1
|
cysteine/tyrosine-rich 1 |
| chr3_+_132316081 | 1.24 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr12_-_91573132 | 1.23 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr2_-_214016314 | 1.23 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr19_+_35939154 | 1.22 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr4_-_159094194 | 1.22 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr2_-_228244013 | 1.22 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr17_+_12453278 | 1.21 |
ENST00000577679.1
|
LINC00670
|
long intergenic non-protein coding RNA 670 |
| chr11_+_131240593 | 1.21 |
ENST00000539799.1
|
NTM
|
neurotrimin |
| chr12_-_7848364 | 1.20 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3 |
| chr10_+_129785536 | 1.20 |
ENST00000419012.2
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr3_+_151451707 | 1.19 |
ENST00000356517.3
|
AADACL2
|
arylacetamide deacetylase-like 2 |
| chr15_+_84841242 | 1.19 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr8_-_95274536 | 1.19 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr1_+_186649754 | 1.19 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr9_-_35691017 | 1.18 |
ENST00000378292.3
|
TPM2
|
tropomyosin 2 (beta) |
| chr14_+_63671577 | 1.18 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr2_-_179914760 | 1.17 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr5_-_94417339 | 1.16 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr1_+_203734296 | 1.16 |
ENST00000442561.2
ENST00000367217.5 |
LAX1
|
lymphocyte transmembrane adaptor 1 |
| chr11_+_118754475 | 1.16 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr10_+_129785574 | 1.14 |
ENST00000430713.2
ENST00000471218.1 |
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr14_-_92413727 | 1.14 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr2_-_42160486 | 1.14 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr11_-_102651343 | 1.12 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr4_+_86749045 | 1.11 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr11_-_64660916 | 1.11 |
ENST00000413053.1
|
MIR194-2
|
microRNA 194-2 |
| chr6_-_112081113 | 1.11 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr18_+_61575200 | 1.11 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr6_-_108145499 | 1.11 |
ENST00000369020.3
ENST00000369022.2 |
SCML4
|
sex comb on midleg-like 4 (Drosophila) |
| chr1_+_66820058 | 1.10 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr12_-_16759711 | 1.09 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr18_-_22932080 | 1.09 |
ENST00000584787.1
ENST00000361524.3 ENST00000538137.2 |
ZNF521
|
zinc finger protein 521 |
| chr8_-_131399110 | 1.09 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr2_+_189839046 | 1.09 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr15_+_63354769 | 1.08 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr19_-_55150343 | 1.08 |
ENST00000456337.1
|
AC009892.10
|
Uncharacterized protein |
| chr1_-_85462762 | 1.08 |
ENST00000284027.5
|
MCOLN2
|
mucolipin 2 |
| chr9_-_215744 | 1.07 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr10_-_105212059 | 1.07 |
ENST00000260743.5
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr6_+_106535455 | 1.06 |
ENST00000424894.1
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr1_+_175036966 | 1.05 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr12_-_91573249 | 1.05 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr3_+_151986709 | 1.05 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr1_-_168513229 | 1.05 |
ENST00000367819.2
|
XCL2
|
chemokine (C motif) ligand 2 |
| chr10_+_11206925 | 1.04 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr9_-_16870704 | 1.04 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr14_+_22475742 | 1.04 |
ENST00000390447.3
|
TRAV19
|
T cell receptor alpha variable 19 |
| chr12_-_48226897 | 1.03 |
ENST00000434070.1
|
HDAC7
|
histone deacetylase 7 |
| chr9_-_136006496 | 1.03 |
ENST00000372062.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chrX_-_129244655 | 1.02 |
ENST00000335997.7
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr18_+_77155942 | 1.01 |
ENST00000397790.2
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr8_+_95653302 | 1.01 |
ENST00000423620.2
ENST00000433389.2 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr12_-_16760021 | 1.00 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr8_+_95653427 | 1.00 |
ENST00000454170.2
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr1_+_22351977 | 1.00 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr3_+_69812877 | 1.00 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr1_+_215179188 | 0.99 |
ENST00000391895.2
|
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr17_-_34207295 | 0.99 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr11_+_392587 | 0.99 |
ENST00000534401.1
|
PKP3
|
plakophilin 3 |
| chr8_-_30002179 | 0.98 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr4_+_41614909 | 0.98 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr16_-_21436459 | 0.98 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr7_+_28725585 | 0.97 |
ENST00000396298.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr19_+_38779778 | 0.97 |
ENST00000590738.1
ENST00000587519.2 ENST00000591889.1 |
SPINT2
CTB-102L5.4
|
serine peptidase inhibitor, Kunitz type, 2 CTB-102L5.4 |
| chr10_-_105212141 | 0.97 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr4_+_154387480 | 0.96 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr8_+_95653373 | 0.96 |
ENST00000358397.5
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr8_+_70404996 | 0.96 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr14_+_75761099 | 0.95 |
ENST00000561000.1
ENST00000558575.1 |
RP11-293M10.5
|
RP11-293M10.5 |
| chr1_-_85462623 | 0.94 |
ENST00000370608.3
|
MCOLN2
|
mucolipin 2 |
| chr5_+_38445641 | 0.94 |
ENST00000397210.3
ENST00000506135.1 ENST00000508131.1 |
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains |
| chr12_-_16759440 | 0.94 |
ENST00000537304.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_+_177159744 | 0.93 |
ENST00000439009.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr4_-_41750922 | 0.93 |
ENST00000226382.2
|
PHOX2B
|
paired-like homeobox 2b |
| chr3_-_71353892 | 0.92 |
ENST00000484350.1
|
FOXP1
|
forkhead box P1 |
| chr12_-_91573316 | 0.91 |
ENST00000393155.1
|
DCN
|
decorin |
| chr2_+_28618532 | 0.89 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr1_+_84873913 | 0.88 |
ENST00000370662.3
|
DNASE2B
|
deoxyribonuclease II beta |
| chr3_+_177159695 | 0.88 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr12_-_52685312 | 0.88 |
ENST00000327741.5
|
KRT81
|
keratin 81 |
| chrX_-_19905577 | 0.87 |
ENST00000379697.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr16_+_68573116 | 0.87 |
ENST00000570495.1
ENST00000563169.2 ENST00000564323.1 ENST00000562156.1 ENST00000573685.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr4_-_80994471 | 0.87 |
ENST00000295465.4
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr8_-_122653630 | 0.86 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr12_-_10605929 | 0.86 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr11_-_58612168 | 0.84 |
ENST00000287275.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr3_+_69134080 | 0.83 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr14_-_54418598 | 0.83 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr3_-_50540854 | 0.82 |
ENST00000423994.2
ENST00000424201.2 ENST00000479441.1 ENST00000429770.1 |
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr9_+_27109133 | 0.81 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 1.0 | 3.8 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 1.0 | 3.8 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 1.0 | 4.8 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.9 | 2.8 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.9 | 5.2 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.8 | 2.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.8 | 3.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.7 | 2.0 | GO:2000409 | astrocyte chemotaxis(GO:0035700) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) positive regulation of T cell extravasation(GO:2000409) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.6 | 1.9 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.6 | 2.8 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.5 | 4.4 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.5 | 1.6 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 0.5 | 2.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.4 | 2.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.4 | 1.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.4 | 1.9 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.4 | 4.5 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.4 | 8.0 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.3 | 3.4 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.3 | 1.6 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.3 | 0.9 | GO:0021934 | medulla oblongata development(GO:0021550) hindbrain tangential cell migration(GO:0021934) lateral line system development(GO:0048925) |
| 0.3 | 1.2 | GO:1905229 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.3 | 1.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 2.1 | GO:1901526 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.3 | 2.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.3 | 1.4 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.3 | 2.2 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.3 | 2.5 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 0.8 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.3 | 1.6 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 3.4 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.3 | 3.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.3 | 0.8 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.2 | 1.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 2.9 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 1.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 | 0.9 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 3.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 1.0 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.2 | 2.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 1.6 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.2 | 1.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 0.7 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 1.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.2 | 0.7 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.2 | 2.8 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.2 | 0.7 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.2 | 1.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 0.6 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 0.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.8 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 1.9 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 0.6 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.2 | 7.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 1.6 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.4 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 1.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 2.0 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 1.4 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 2.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 1.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 2.6 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 2.0 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.4 | GO:2000563 | positive regulation of thymocyte migration(GO:2000412) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 2.6 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.1 | 0.5 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.1 | 1.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 2.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 3.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 2.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.6 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.5 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 2.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.2 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.7 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.3 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 1.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.5 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 1.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.7 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 1.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.9 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.3 | GO:0036315 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) |
| 0.1 | 5.9 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 1.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.1 | 1.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.6 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 1.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.4 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 2.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.7 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.8 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.1 | 0.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.3 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 3.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.7 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.0 | 0.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 1.3 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.2 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.5 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 2.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:2000330 | interleukin-23-mediated signaling pathway(GO:0038155) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.6 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.4 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 1.7 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 2.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.3 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 2.7 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 1.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 2.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.4 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.5 | GO:0090101 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway(GO:0090101) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 6.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 1.5 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 5.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 2.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.9 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 4.5 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 5.3 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 1.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.8 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.8 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 1.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.8 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 1.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 2.9 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.9 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.5 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 1.0 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.5 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 4.7 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.9 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.5 | GO:0048679 | activation of MAPKKK activity(GO:0000185) regulation of axon regeneration(GO:0048679) |
| 0.0 | 0.8 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.7 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.7 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.0 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.7 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.5 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0097679 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) other organism cytoplasm(GO:0097679) |
| 0.3 | 2.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 3.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 2.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 3.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 6.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 7.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 0.7 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.4 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 3.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 4.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 3.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 1.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.7 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 4.8 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 3.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 2.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.0 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.2 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.1 | 1.2 | GO:0042995 | cell projection(GO:0042995) |
| 0.1 | 11.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 4.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 16.1 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 6.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 8.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 1.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.6 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.8 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.9 | 3.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.9 | 2.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.6 | 3.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 1.6 | GO:0030395 | lactose binding(GO:0030395) |
| 0.5 | 3.4 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.5 | 2.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.5 | 1.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 7.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.4 | 2.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 1.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.4 | 4.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.4 | 2.8 | GO:0042806 | fucose binding(GO:0042806) |
| 0.4 | 3.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 2.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 0.9 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.3 | 6.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 6.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 1.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.2 | 2.5 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.2 | 4.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 1.7 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.2 | 2.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 2.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 0.6 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.2 | 1.6 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.2 | 3.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 1.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.2 | 0.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 0.8 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.9 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 1.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 2.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 1.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 1.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 4.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.8 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 1.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.6 | GO:0052846 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 4.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 1.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 8.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 3.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 7.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 1.7 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.1 | 2.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.3 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 1.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.5 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 1.2 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.2 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 2.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 3.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 1.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 1.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 2.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.1 | GO:0008266 | poly-pyrimidine tract binding(GO:0008187) poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.4 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 1.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 1.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 2.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 3.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.7 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 6.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 3.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 1.5 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.3 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.5 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 3.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 8.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 1.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 4.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 9.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 3.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 6.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 3.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 5.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 6.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 4.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 3.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 7.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 12.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 4.5 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 1.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 6.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 2.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 1.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.0 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.9 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 3.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 2.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.8 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 5.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.7 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.2 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |