Project
Illumina Body Map 2
Navigation
Downloads
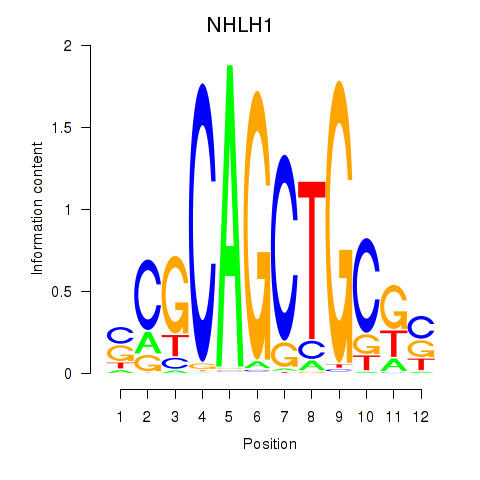
Results for NHLH1
Z-value: 1.50
Transcription factors associated with NHLH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHLH1
|
ENSG00000171786.5 | nescient helix-loop-helix 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NHLH1 | hg19_v2_chr1_+_160336851_160336868 | 0.37 | 3.7e-02 | Click! |
Activity profile of NHLH1 motif
Sorted Z-values of NHLH1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_105036909 | 4.93 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr11_+_17756279 | 4.87 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr11_-_35441597 | 4.54 |
ENST00000395753.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr11_-_35441524 | 4.47 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr21_+_34442439 | 3.89 |
ENST00000382348.1
ENST00000333063.5 |
OLIG1
|
oligodendrocyte transcription factor 1 |
| chr16_+_23847339 | 3.78 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr8_+_24771265 | 3.76 |
ENST00000518131.1
ENST00000437366.2 |
NEFM
|
neurofilament, medium polypeptide |
| chr16_+_23847267 | 3.73 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr8_+_24772455 | 3.73 |
ENST00000433454.2
|
NEFM
|
neurofilament, medium polypeptide |
| chr2_-_11810284 | 3.69 |
ENST00000306928.5
|
NTSR2
|
neurotensin receptor 2 |
| chr16_+_330581 | 3.28 |
ENST00000219409.3
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr1_-_21948906 | 3.27 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr3_-_62860878 | 3.20 |
ENST00000283269.9
|
CADPS
|
Ca++-dependent secretion activator |
| chrX_-_43832711 | 3.01 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr12_-_103352144 | 2.98 |
ENST00000551337.1
|
PAH
|
phenylalanine hydroxylase |
| chr1_+_33352036 | 2.71 |
ENST00000373467.3
|
HPCA
|
hippocalcin |
| chr8_-_139509065 | 2.56 |
ENST00000395297.1
|
FAM135B
|
family with sequence similarity 135, member B |
| chr1_-_193155729 | 2.55 |
ENST00000367434.4
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr2_+_105471969 | 2.54 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr6_-_84419101 | 2.54 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr8_-_33455268 | 2.54 |
ENST00000522982.1
|
DUSP26
|
dual specificity phosphatase 26 (putative) |
| chr5_+_126626498 | 2.53 |
ENST00000503335.2
ENST00000508365.1 ENST00000418761.2 ENST00000274473.6 |
MEGF10
|
multiple EGF-like-domains 10 |
| chr2_-_102003987 | 2.52 |
ENST00000324768.5
|
CREG2
|
cellular repressor of E1A-stimulated genes 2 |
| chr9_-_23825956 | 2.51 |
ENST00000397312.2
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr6_-_84418841 | 2.45 |
ENST00000369694.2
ENST00000195649.6 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr11_+_125774258 | 2.44 |
ENST00000263576.6
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr4_-_168155300 | 2.40 |
ENST00000541637.1
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_-_168155577 | 2.39 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_-_142682178 | 2.35 |
ENST00000340634.3
|
PAQR9
|
progestin and adipoQ receptor family member IX |
| chr10_+_103348031 | 2.30 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr4_-_168155169 | 2.29 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr19_+_12949251 | 2.29 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr4_-_186696561 | 2.26 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr20_+_20348740 | 2.24 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr12_-_118406777 | 2.23 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr3_-_47619623 | 2.23 |
ENST00000456150.1
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C) |
| chr9_-_23826298 | 2.21 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr6_-_84418860 | 2.20 |
ENST00000521743.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr4_-_168155700 | 2.20 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_-_183146369 | 2.16 |
ENST00000482017.1
|
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr11_+_7273181 | 2.16 |
ENST00000318881.6
|
SYT9
|
synaptotagmin IX |
| chr2_+_231902193 | 2.13 |
ENST00000373640.4
|
C2orf72
|
chromosome 2 open reading frame 72 |
| chr16_+_23847355 | 2.12 |
ENST00000498058.1
|
PRKCB
|
protein kinase C, beta |
| chr5_+_167181917 | 2.12 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr19_+_4304685 | 2.11 |
ENST00000601006.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr3_+_96533413 | 2.11 |
ENST00000470610.2
ENST00000389672.5 |
EPHA6
|
EPH receptor A6 |
| chr4_-_168155730 | 2.11 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_-_186696515 | 2.09 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_29624898 | 2.06 |
ENST00000396704.3
ENST00000483013.1 ENST00000490427.1 ENST00000416766.2 ENST00000376891.4 ENST00000376898.3 ENST00000396701.2 ENST00000494692.1 ENST00000431798.2 |
MOG
|
myelin oligodendrocyte glycoprotein |
| chr7_-_100808843 | 2.06 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr1_-_109849612 | 2.06 |
ENST00000357155.1
|
MYBPHL
|
myosin binding protein H-like |
| chrX_+_110187513 | 2.06 |
ENST00000446737.1
ENST00000425146.1 |
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr11_+_107461804 | 2.04 |
ENST00000531234.1
|
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr4_+_89300158 | 2.04 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr3_-_133614421 | 2.03 |
ENST00000543906.1
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr1_+_50575292 | 2.03 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr4_+_154074217 | 2.01 |
ENST00000437508.2
|
TRIM2
|
tripartite motif containing 2 |
| chr11_-_40315640 | 1.98 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr19_+_4304585 | 1.97 |
ENST00000221856.6
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr6_+_72596604 | 1.96 |
ENST00000348717.5
ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr11_+_71249071 | 1.95 |
ENST00000398534.3
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr19_-_17559376 | 1.94 |
ENST00000341130.5
|
TMEM221
|
transmembrane protein 221 |
| chrX_+_84498989 | 1.93 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chr19_+_4304632 | 1.93 |
ENST00000597590.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr1_+_177140633 | 1.89 |
ENST00000361539.4
|
BRINP2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr7_+_20370300 | 1.88 |
ENST00000537992.1
|
ITGB8
|
integrin, beta 8 |
| chr16_-_74808710 | 1.87 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr12_-_86650045 | 1.87 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr12_+_27932803 | 1.84 |
ENST00000381271.2
|
KLHL42
|
kelch-like family member 42 |
| chr19_-_55953704 | 1.83 |
ENST00000416792.1
|
SHISA7
|
shisa family member 7 |
| chr4_-_186696425 | 1.82 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_+_100150622 | 1.82 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr16_+_6533729 | 1.81 |
ENST00000551752.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chrX_+_84499081 | 1.80 |
ENST00000276123.3
|
ZNF711
|
zinc finger protein 711 |
| chr19_+_39989580 | 1.80 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chrX_+_84499038 | 1.79 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chr4_-_168155417 | 1.79 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr11_+_71238313 | 1.78 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr14_-_45603657 | 1.77 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr12_-_6580094 | 1.76 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr20_-_61992738 | 1.74 |
ENST00000370263.4
|
CHRNA4
|
cholinergic receptor, nicotinic, alpha 4 (neuronal) |
| chr12_-_6579833 | 1.74 |
ENST00000396308.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr1_-_85156090 | 1.74 |
ENST00000605755.1
ENST00000437941.2 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr13_+_88324870 | 1.74 |
ENST00000325089.6
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr1_+_202317855 | 1.74 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr17_-_79359046 | 1.73 |
ENST00000574041.1
|
RP11-1055B8.3
|
RP11-1055B8.3 |
| chr17_-_31620006 | 1.72 |
ENST00000225823.2
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr15_-_102264619 | 1.72 |
ENST00000335968.3
|
TARSL2
|
threonyl-tRNA synthetase-like 2 |
| chr3_-_133614597 | 1.71 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr3_+_96533621 | 1.70 |
ENST00000542517.1
ENST00000506569.1 |
EPHA6
|
EPH receptor A6 |
| chr4_-_186696636 | 1.70 |
ENST00000444771.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_167182003 | 1.69 |
ENST00000520394.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr1_-_85156417 | 1.69 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr4_-_139163491 | 1.69 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr17_-_79359154 | 1.68 |
ENST00000572077.1
|
RP11-1055B8.3
|
RP11-1055B8.3 |
| chr1_-_85156216 | 1.68 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr1_-_33336414 | 1.68 |
ENST00000373471.3
ENST00000609187.1 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr10_-_135379132 | 1.68 |
ENST00000343131.5
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chr6_-_84418738 | 1.67 |
ENST00000519779.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr17_+_80693427 | 1.67 |
ENST00000300784.7
|
FN3K
|
fructosamine 3 kinase |
| chr5_+_176237478 | 1.64 |
ENST00000329542.4
|
UNC5A
|
unc-5 homolog A (C. elegans) |
| chr1_-_177133818 | 1.63 |
ENST00000424564.2
ENST00000361833.2 |
ASTN1
|
astrotactin 1 |
| chr12_+_121088291 | 1.62 |
ENST00000351200.2
|
CABP1
|
calcium binding protein 1 |
| chr7_-_766879 | 1.62 |
ENST00000537384.1
ENST00000417852.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr4_+_30723003 | 1.62 |
ENST00000543491.1
|
PCDH7
|
protocadherin 7 |
| chr19_-_55954230 | 1.61 |
ENST00000376325.4
|
SHISA7
|
shisa family member 7 |
| chr16_+_6533380 | 1.61 |
ENST00000552089.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr17_-_79359021 | 1.60 |
ENST00000572301.1
|
RP11-1055B8.3
|
RP11-1055B8.3 |
| chr5_-_19988339 | 1.60 |
ENST00000382275.1
|
CDH18
|
cadherin 18, type 2 |
| chr12_-_86650154 | 1.59 |
ENST00000552435.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr10_+_18689637 | 1.59 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr10_+_18429671 | 1.58 |
ENST00000282343.8
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr20_-_43438912 | 1.56 |
ENST00000541604.2
ENST00000372851.3 |
RIMS4
|
regulating synaptic membrane exocytosis 4 |
| chr2_+_11052054 | 1.55 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr1_-_177134024 | 1.55 |
ENST00000367654.3
|
ASTN1
|
astrotactin 1 |
| chr5_-_19988288 | 1.54 |
ENST00000502796.1
ENST00000511273.1 |
CDH18
|
cadherin 18, type 2 |
| chr12_-_86650077 | 1.54 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr20_+_58179582 | 1.52 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr4_-_83719983 | 1.51 |
ENST00000319540.4
|
SCD5
|
stearoyl-CoA desaturase 5 |
| chr16_+_7560114 | 1.51 |
ENST00000570626.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr17_+_74864476 | 1.51 |
ENST00000301618.4
ENST00000569840.2 |
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr7_-_100808394 | 1.50 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr4_+_176987131 | 1.50 |
ENST00000280190.4
|
WDR17
|
WD repeat domain 17 |
| chr2_-_232791038 | 1.50 |
ENST00000295440.2
ENST00000409852.1 |
NPPC
|
natriuretic peptide C |
| chr3_+_50712672 | 1.50 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr12_+_110152033 | 1.49 |
ENST00000538780.1
|
FAM222A
|
family with sequence similarity 222, member A |
| chr6_+_29624862 | 1.49 |
ENST00000376894.4
|
MOG
|
myelin oligodendrocyte glycoprotein |
| chr19_-_49944806 | 1.49 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr3_-_68981685 | 1.49 |
ENST00000495737.1
ENST00000295569.7 |
FAM19A4
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4 |
| chr15_+_40650408 | 1.49 |
ENST00000267889.3
|
DISP2
|
dispatched homolog 2 (Drosophila) |
| chr9_+_37650945 | 1.48 |
ENST00000377765.3
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr6_+_150464155 | 1.47 |
ENST00000361131.4
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr16_-_29910365 | 1.47 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr14_+_70346125 | 1.46 |
ENST00000361956.3
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr7_-_767249 | 1.44 |
ENST00000403562.1
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr22_+_19705928 | 1.44 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr1_-_47134101 | 1.43 |
ENST00000576409.1
|
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr12_-_67197760 | 1.42 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr2_+_16080659 | 1.41 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chrX_+_12156582 | 1.41 |
ENST00000380682.1
|
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr22_-_37823468 | 1.40 |
ENST00000402918.2
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr12_+_110172572 | 1.40 |
ENST00000358906.3
|
FAM222A
|
family with sequence similarity 222, member A |
| chr9_-_102582155 | 1.39 |
ENST00000427039.1
|
RP11-554F20.1
|
RP11-554F20.1 |
| chr13_+_88325498 | 1.39 |
ENST00000400028.3
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr14_+_62584197 | 1.38 |
ENST00000334389.4
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr19_-_51220176 | 1.38 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr3_-_133614467 | 1.38 |
ENST00000469959.1
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr1_-_85155939 | 1.37 |
ENST00000603677.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr6_+_29624758 | 1.36 |
ENST00000376917.3
ENST00000376902.3 ENST00000533330.2 ENST00000376888.2 |
MOG
|
myelin oligodendrocyte glycoprotein |
| chr4_+_184826418 | 1.36 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr9_-_111929560 | 1.34 |
ENST00000561981.2
|
FRRS1L
|
ferric-chelate reductase 1-like |
| chr4_+_176986978 | 1.34 |
ENST00000508596.1
ENST00000393643.2 |
WDR17
|
WD repeat domain 17 |
| chr2_-_86564776 | 1.34 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr12_+_56075330 | 1.33 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr4_+_2061119 | 1.33 |
ENST00000423729.2
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative) |
| chr19_+_39989535 | 1.32 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr12_-_74686314 | 1.30 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr1_+_2065077 | 1.30 |
ENST00000471018.2
|
PRKCZ
|
protein kinase C, zeta |
| chr20_-_9819479 | 1.29 |
ENST00000378423.1
ENST00000353224.5 |
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr13_-_96296944 | 1.29 |
ENST00000361396.2
ENST00000376829.2 |
DZIP1
|
DAZ interacting zinc finger protein 1 |
| chr12_+_50344516 | 1.28 |
ENST00000199280.3
ENST00000550862.1 |
AQP2
|
aquaporin 2 (collecting duct) |
| chr7_+_45613958 | 1.28 |
ENST00000297323.7
|
ADCY1
|
adenylate cyclase 1 (brain) |
| chr1_+_15272271 | 1.28 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr3_+_167453026 | 1.27 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr1_-_177133998 | 1.27 |
ENST00000367657.3
|
ASTN1
|
astrotactin 1 |
| chr15_+_43803143 | 1.27 |
ENST00000382031.1
|
MAP1A
|
microtubule-associated protein 1A |
| chr6_+_146056706 | 1.26 |
ENST00000603994.1
|
RP3-466P17.1
|
RP3-466P17.1 |
| chr6_-_80657292 | 1.25 |
ENST00000369816.4
|
ELOVL4
|
ELOVL fatty acid elongase 4 |
| chr1_+_233749739 | 1.25 |
ENST00000366621.3
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr4_-_186697044 | 1.25 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_181160240 | 1.23 |
ENST00000460993.1
|
RP11-275H4.1
|
RP11-275H4.1 |
| chr2_-_73511559 | 1.23 |
ENST00000521871.1
|
FBXO41
|
F-box protein 41 |
| chr10_-_98945677 | 1.23 |
ENST00000266058.4
ENST00000371041.3 |
SLIT1
|
slit homolog 1 (Drosophila) |
| chr1_-_85930823 | 1.23 |
ENST00000284031.8
ENST00000539042.1 |
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr14_-_65438865 | 1.22 |
ENST00000267512.5
|
RAB15
|
RAB15, member RAS oncogene family |
| chr11_-_71293921 | 1.22 |
ENST00000398530.1
|
KRTAP5-11
|
keratin associated protein 5-11 |
| chr2_+_103236004 | 1.22 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chrX_+_49126294 | 1.21 |
ENST00000466508.1
ENST00000438316.1 ENST00000055335.6 ENST00000495799.1 |
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F |
| chr1_-_237167718 | 1.20 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chr5_+_113697983 | 1.19 |
ENST00000264773.3
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr3_+_35681081 | 1.19 |
ENST00000428373.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr16_+_330448 | 1.18 |
ENST00000447871.1
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr12_-_6579808 | 1.18 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr17_-_56606705 | 1.18 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr3_-_172428842 | 1.18 |
ENST00000424772.1
|
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr20_-_9819674 | 1.17 |
ENST00000378429.3
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr1_+_151693984 | 1.16 |
ENST00000479191.1
|
RIIAD1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 |
| chr3_-_133614297 | 1.16 |
ENST00000486858.1
ENST00000477759.1 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr11_-_1619524 | 1.16 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr1_-_84464780 | 1.16 |
ENST00000260505.8
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7 |
| chr6_+_72596406 | 1.15 |
ENST00000491071.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr13_+_35516390 | 1.15 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr18_+_43913919 | 1.14 |
ENST00000587853.1
|
RNF165
|
ring finger protein 165 |
| chr3_+_159481791 | 1.14 |
ENST00000460298.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr16_+_90086002 | 1.14 |
ENST00000563936.1
ENST00000536122.1 ENST00000561675.1 |
GAS8
|
growth arrest-specific 8 |
| chr3_-_62860704 | 1.14 |
ENST00000490353.2
|
CADPS
|
Ca++-dependent secretion activator |
| chr19_+_14184370 | 1.14 |
ENST00000590772.1
|
hsa-mir-1199
|
hsa-mir-1199 |
| chr1_-_38218577 | 1.13 |
ENST00000540011.1
|
EPHA10
|
EPH receptor A10 |
| chr8_-_17270809 | 1.13 |
ENST00000180173.5
ENST00000521857.1 |
MTMR7
|
myotubularin related protein 7 |
| chr13_-_36705425 | 1.13 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NHLH1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 2.4 | 9.6 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 1.1 | 3.3 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.8 | 9.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.6 | 3.2 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.6 | 2.5 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.6 | 4.3 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.6 | 2.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.5 | 13.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.5 | 4.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.5 | 2.7 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.5 | 1.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.5 | 3.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.5 | 2.5 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.5 | 1.5 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.5 | 3.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.5 | 1.5 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.5 | 1.5 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.5 | 2.4 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.5 | 0.5 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.5 | 1.9 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.5 | 1.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.5 | 1.4 | GO:0050894 | determination of affect(GO:0050894) |
| 0.4 | 1.7 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.4 | 4.9 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.4 | 1.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.4 | 3.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.4 | 3.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.4 | 2.7 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.4 | 2.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 1.1 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.3 | 1.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.3 | 1.9 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.3 | 1.3 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.3 | 1.4 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.3 | 2.5 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) recognition of apoptotic cell(GO:0043654) |
| 0.3 | 6.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.3 | 0.8 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.2 | 1.7 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 0.7 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.2 | 1.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.2 | 1.4 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 0.9 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 1.5 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 1.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 9.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 0.8 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.2 | 2.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.4 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 1.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 0.6 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.2 | 2.3 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.2 | 0.7 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.2 | 4.7 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 8.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 0.7 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.2 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.2 | 3.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 3.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 2.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 1.9 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 3.0 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.4 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 3.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 1.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 1.7 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.6 | GO:0018963 | insecticide metabolic process(GO:0017143) phthalate metabolic process(GO:0018963) |
| 0.1 | 0.9 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 1.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.1 | 0.4 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 1.8 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 3.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 3.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 1.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 2.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.8 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 | 2.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.5 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.3 | GO:0001933 | negative regulation of protein phosphorylation(GO:0001933) |
| 0.1 | 1.5 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 1.3 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.1 | 3.1 | GO:0021756 | striatum development(GO:0021756) |
| 0.1 | 1.0 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 4.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 1.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 1.7 | GO:0030432 | peristalsis(GO:0030432) |
| 0.1 | 1.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.6 | GO:0015853 | adenine transport(GO:0015853) |
| 0.1 | 1.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.6 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.1 | 0.8 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 0.8 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.5 | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 3.4 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 1.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 1.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.3 | GO:1902308 | regulation of peptidyl-serine dephosphorylation(GO:1902308) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 1.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 1.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 3.0 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 6.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 2.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 1.3 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.2 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 1.2 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 1.5 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 1.8 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.1 | 1.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.4 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.1 | 0.9 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.7 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 1.0 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.9 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 1.6 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 3.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.9 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 1.0 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) |
| 0.0 | 1.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 3.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 3.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.4 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.8 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 1.6 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.5 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 2.5 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.6 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 2.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 1.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.4 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 1.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.5 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.4 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 1.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 6.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.0 | 0.6 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 3.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.7 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 1.3 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.1 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 0.7 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.8 | GO:0016050 | vesicle organization(GO:0016050) |
| 0.0 | 1.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.3 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 0.4 | GO:0031214 | biomineral tissue development(GO:0031214) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.4 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.2 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 2.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 3.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.7 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.6 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 1.6 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.4 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.3 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.4 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 1.7 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.0 | 0.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 1.0 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.3 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.5 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.4 | GO:0021696 | cerebellar cortex morphogenesis(GO:0021696) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 2.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.3 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.3 | GO:0051197 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 3.7 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 4.8 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.0 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.4 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 12.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.6 | 1.9 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.3 | 0.6 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.3 | 0.8 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 1.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 12.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 2.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 1.7 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 2.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.0 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.2 | 6.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.5 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.1 | 2.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 3.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.8 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 2.0 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 0.5 | GO:0032279 | axonemal microtubule(GO:0005879) asymmetric synapse(GO:0032279) |
| 0.1 | 1.6 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 2.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 4.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 3.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 2.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.6 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 8.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 3.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.8 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 4.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.4 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 1.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 8.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.3 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 14.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 5.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.6 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 7.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 4.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 5.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.8 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 2.0 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 3.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 3.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 3.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 6.1 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 2.8 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 3.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0000779 | condensed chromosome kinetochore(GO:0000777) condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 2.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 9.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.9 | 9.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.9 | 2.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.6 | 5.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.5 | 3.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.5 | 3.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.5 | 1.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.5 | 1.4 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.4 | 1.7 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.4 | 13.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 2.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.4 | 1.6 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 3.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.4 | 4.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.7 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.3 | 1.9 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.3 | 1.6 | GO:0033265 | choline binding(GO:0033265) |
| 0.3 | 2.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 1.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 3.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 1.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.3 | 2.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.3 | 0.8 | GO:0035643 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.3 | 1.5 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 1.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 1.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 9.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 9.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 3.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.7 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 6.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 1.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 0.2 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.2 | 0.9 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.2 | 1.5 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 1.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 1.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.2 | 0.6 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.2 | 4.9 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 5.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.5 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 1.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 0.4 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.1 | 3.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 4.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 2.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.9 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 1.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 1.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 2.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 3.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 1.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 3.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.6 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 4.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 2.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 2.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 3.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.8 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 2.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.7 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 2.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.2 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 1.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 10.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 1.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.9 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 1.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.3 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 3.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.6 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 2.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 4.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.6 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 2.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 6.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.5 | GO:0016462 | pyrophosphatase activity(GO:0016462) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 2.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 1.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 9.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 4.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 3.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 5.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 9.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.2 | 3.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 2.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 5.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 2.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 10.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 6.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 4.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 4.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 2.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 4.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 2.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.4 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 1.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |