Project
Illumina Body Map 2
Navigation
Downloads
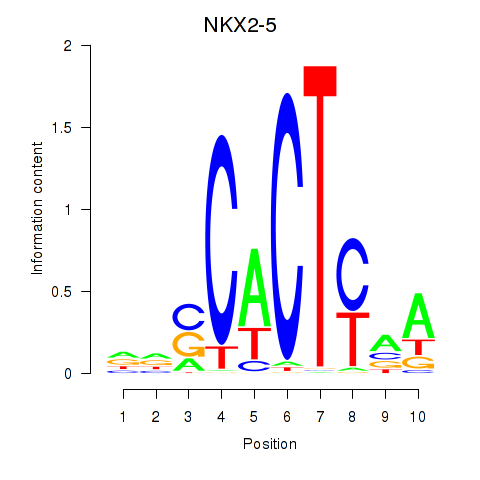
Results for NKX2-5
Z-value: 1.11
Transcription factors associated with NKX2-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-5
|
ENSG00000183072.9 | NK2 homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-5 | hg19_v2_chr5_-_172662303_172662360 | -0.11 | 5.5e-01 | Click! |
Activity profile of NKX2-5 motif
Sorted Z-values of NKX2-5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_81320151 | 8.31 |
ENST00000372325.2
ENST00000372327.5 ENST00000417041.1 |
SFTPA2
|
surfactant protein A2 |
| chr14_+_22356029 | 5.14 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr14_+_22309368 | 4.88 |
ENST00000390433.1
|
TRAV12-1
|
T cell receptor alpha variable 12-1 |
| chr10_+_81370689 | 4.71 |
ENST00000372308.3
ENST00000398636.3 ENST00000428376.2 ENST00000372313.5 ENST00000419470.2 ENST00000429958.1 ENST00000439264.1 |
SFTPA1
|
surfactant protein A1 |
| chr16_+_32859034 | 4.22 |
ENST00000567458.2
ENST00000560724.1 |
IGHV2OR16-5
|
immunoglobulin heavy variable 2/OR16-5 (non-functional) |
| chr14_-_106758101 | 3.78 |
ENST00000390611.2
|
IGHV2-26
|
immunoglobulin heavy variable 2-26 |
| chr4_-_74853897 | 3.49 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr14_+_22788560 | 3.47 |
ENST00000390468.1
|
TRAV41
|
T cell receptor alpha variable 41 |
| chr9_-_117692697 | 3.31 |
ENST00000223795.2
|
TNFSF8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr1_-_36947652 | 3.26 |
ENST00000533491.1
|
CSF3R
|
colony stimulating factor 3 receptor (granulocyte) |
| chr14_-_107179265 | 3.24 |
ENST00000390634.2
|
IGHV2-70
|
immunoglobulin heavy variable 2-70 |
| chr1_+_161676739 | 3.15 |
ENST00000236938.6
ENST00000367959.2 ENST00000546024.1 ENST00000540521.1 ENST00000367949.2 ENST00000350710.3 ENST00000540926.1 |
FCRLA
|
Fc receptor-like A |
| chr1_-_36947120 | 3.10 |
ENST00000361632.4
|
CSF3R
|
colony stimulating factor 3 receptor (granulocyte) |
| chr1_-_206306107 | 2.96 |
ENST00000436158.1
ENST00000455672.1 |
RP11-38J22.6
|
RP11-38J22.6 |
| chr14_-_106830057 | 2.86 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34 |
| chr1_-_160616804 | 2.70 |
ENST00000538290.1
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr1_+_161677034 | 2.66 |
ENST00000349527.4
ENST00000309691.6 ENST00000294796.4 ENST00000367953.3 ENST00000367950.1 |
FCRLA
|
Fc receptor-like A |
| chr5_+_147258266 | 2.54 |
ENST00000296694.4
|
SCGB3A2
|
secretoglobin, family 3A, member 2 |
| chr19_+_10947251 | 2.47 |
ENST00000592854.1
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr22_+_40322595 | 2.38 |
ENST00000420971.1
ENST00000544756.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr22_-_50524298 | 2.25 |
ENST00000311597.5
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr22_-_50523760 | 2.12 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr1_+_161676983 | 2.12 |
ENST00000367957.2
|
FCRLA
|
Fc receptor-like A |
| chr22_-_50523807 | 2.08 |
ENST00000442311.1
ENST00000538737.1 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr17_-_29624343 | 2.03 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr15_-_74726283 | 2.03 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr22_-_50523843 | 1.99 |
ENST00000535444.1
ENST00000431262.2 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr22_+_40322623 | 1.95 |
ENST00000399090.2
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr7_+_142919130 | 1.91 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr7_-_24957699 | 1.81 |
ENST00000441059.1
ENST00000415162.1 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr17_-_29641104 | 1.77 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chrX_+_71353499 | 1.77 |
ENST00000373677.1
|
NHSL2
|
NHS-like 2 |
| chr17_-_29641084 | 1.77 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr3_+_45984947 | 1.72 |
ENST00000304552.4
|
CXCR6
|
chemokine (C-X-C motif) receptor 6 |
| chr14_-_106406090 | 1.69 |
ENST00000390593.2
|
IGHV6-1
|
immunoglobulin heavy variable 6-1 |
| chr14_-_106963409 | 1.67 |
ENST00000390621.2
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr19_+_1026566 | 1.67 |
ENST00000348419.3
ENST00000565096.2 ENST00000562958.2 ENST00000562075.2 ENST00000607102.1 |
CNN2
|
calponin 2 |
| chr15_+_81589254 | 1.61 |
ENST00000394652.2
|
IL16
|
interleukin 16 |
| chr2_-_175499294 | 1.61 |
ENST00000392547.2
|
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr14_+_22963806 | 1.60 |
ENST00000390493.1
|
TRAJ44
|
T cell receptor alpha joining 44 |
| chr2_-_85895295 | 1.59 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chr6_-_41715128 | 1.58 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr13_+_95364963 | 1.56 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr16_+_33006369 | 1.45 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr17_-_80291818 | 1.38 |
ENST00000269389.3
ENST00000581691.1 |
SECTM1
|
secreted and transmembrane 1 |
| chr1_+_26606608 | 1.31 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr13_-_95364389 | 1.28 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr5_-_171615315 | 1.27 |
ENST00000176763.5
|
STK10
|
serine/threonine kinase 10 |
| chr4_+_81118647 | 1.27 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8 |
| chr7_-_81399411 | 1.25 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_-_112115103 | 1.21 |
ENST00000462598.3
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr4_-_168155577 | 1.20 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chrX_-_142722897 | 1.18 |
ENST00000338017.4
|
SLITRK4
|
SLIT and NTRK-like family, member 4 |
| chr2_+_152214098 | 1.14 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr7_-_142919360 | 1.13 |
ENST00000595842.1
|
AC073342.1
|
HCG2002387; Uncharacterized protein |
| chr12_+_113354341 | 1.11 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr17_-_79623597 | 1.10 |
ENST00000574024.1
ENST00000331056.5 |
PDE6G
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr11_+_118754475 | 1.08 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr11_-_73689037 | 1.05 |
ENST00000544615.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr1_+_27153173 | 1.00 |
ENST00000374142.4
|
ZDHHC18
|
zinc finger, DHHC-type containing 18 |
| chr5_-_149792295 | 1.00 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr11_+_108093755 | 0.97 |
ENST00000527891.1
ENST00000532931.1 |
ATM
|
ataxia telangiectasia mutated |
| chr5_-_134788086 | 0.97 |
ENST00000537858.1
|
TIFAB
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr7_-_81399355 | 0.96 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr17_+_74261277 | 0.96 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chr4_-_168155730 | 0.95 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr14_+_23002427 | 0.95 |
ENST00000390527.1
|
TRAJ10
|
T cell receptor alpha joining 10 |
| chr12_+_4918342 | 0.95 |
ENST00000280684.3
ENST00000433855.1 |
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6 |
| chr7_-_81399329 | 0.93 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr13_+_24846799 | 0.93 |
ENST00000488060.1
|
SPATA13
|
spermatogenesis associated 13 |
| chr14_-_85705818 | 0.92 |
ENST00000556016.1
|
RP11-789A21.1
|
RP11-789A21.1 |
| chr10_-_128359008 | 0.92 |
ENST00000488181.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr2_+_113403434 | 0.92 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chr17_+_74261413 | 0.90 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr1_+_207494853 | 0.89 |
ENST00000367064.3
ENST00000367063.2 ENST00000391921.4 ENST00000367067.4 ENST00000314754.8 ENST00000367065.5 ENST00000391920.4 ENST00000367062.4 ENST00000343420.6 |
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| chr9_-_20382446 | 0.89 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr17_-_982198 | 0.88 |
ENST00000571945.1
ENST00000536794.2 |
ABR
|
active BCR-related |
| chr3_+_46283863 | 0.88 |
ENST00000545097.1
ENST00000541018.1 |
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr1_+_100817262 | 0.80 |
ENST00000455467.1
|
CDC14A
|
cell division cycle 14A |
| chr1_-_202858227 | 0.79 |
ENST00000367262.3
|
RABIF
|
RAB interacting factor |
| chr4_-_76861392 | 0.78 |
ENST00000505594.1
|
NAAA
|
N-acylethanolamine acid amidase |
| chr2_+_201997492 | 0.77 |
ENST00000494258.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr6_-_112115074 | 0.76 |
ENST00000368667.2
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr15_-_60683326 | 0.75 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chrX_+_150565038 | 0.73 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr11_+_108093839 | 0.72 |
ENST00000452508.2
|
ATM
|
ataxia telangiectasia mutated |
| chr11_-_19223523 | 0.72 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr18_+_61575200 | 0.70 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr13_-_30951282 | 0.69 |
ENST00000420219.1
|
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr19_+_5681153 | 0.68 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr7_-_81399287 | 0.67 |
ENST00000354224.6
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr8_-_73793975 | 0.67 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr7_-_81399438 | 0.67 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chrX_-_13835147 | 0.66 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr1_-_248738080 | 0.66 |
ENST00000328782.2
|
OR2T34
|
olfactory receptor, family 2, subfamily T, member 34 |
| chr5_-_88180342 | 0.65 |
ENST00000502983.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr2_-_201936302 | 0.65 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr7_-_37488834 | 0.64 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr3_-_111314230 | 0.63 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr7_-_107883678 | 0.61 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr3_-_187388173 | 0.61 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr4_+_156680518 | 0.60 |
ENST00000513437.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr17_+_75447326 | 0.60 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr17_-_19771216 | 0.60 |
ENST00000395544.4
|
ULK2
|
unc-51 like autophagy activating kinase 2 |
| chr2_+_48541776 | 0.59 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr17_+_45973516 | 0.58 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr3_+_46283916 | 0.56 |
ENST00000395940.2
|
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr7_-_37488777 | 0.56 |
ENST00000445322.1
ENST00000448602.1 |
ELMO1
|
engulfment and cell motility 1 |
| chr14_+_81421710 | 0.56 |
ENST00000342443.6
|
TSHR
|
thyroid stimulating hormone receptor |
| chr1_+_76540386 | 0.55 |
ENST00000328299.3
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr7_+_129007964 | 0.55 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr4_-_168155700 | 0.54 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr7_+_129906660 | 0.53 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr10_-_128359074 | 0.52 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr1_+_114472222 | 0.49 |
ENST00000369558.1
ENST00000369561.4 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr17_+_29421987 | 0.47 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr3_+_186692745 | 0.47 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr14_+_64565442 | 0.47 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr18_+_13382553 | 0.47 |
ENST00000586222.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr20_+_57226284 | 0.46 |
ENST00000458280.1
ENST00000355957.5 ENST00000361770.5 ENST00000312283.8 ENST00000412911.1 ENST00000359617.4 ENST00000371141.4 |
STX16
|
syntaxin 16 |
| chr14_+_81421921 | 0.45 |
ENST00000554263.1
ENST00000554435.1 |
TSHR
|
thyroid stimulating hormone receptor |
| chr12_-_49259643 | 0.44 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr3_-_15374659 | 0.44 |
ENST00000426925.1
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr2_-_96874553 | 0.43 |
ENST00000337288.5
ENST00000443962.1 |
STARD7
|
StAR-related lipid transfer (START) domain containing 7 |
| chr5_+_55147205 | 0.43 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr4_-_46126093 | 0.43 |
ENST00000295452.4
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr4_+_2470664 | 0.41 |
ENST00000314289.8
ENST00000541204.1 ENST00000502316.1 ENST00000507247.1 ENST00000509258.1 ENST00000511859.1 |
RNF4
|
ring finger protein 4 |
| chr17_+_58755184 | 0.41 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr1_-_102312600 | 0.40 |
ENST00000359814.3
|
OLFM3
|
olfactomedin 3 |
| chr14_+_81421861 | 0.40 |
ENST00000298171.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr9_+_127020202 | 0.39 |
ENST00000373600.3
ENST00000320246.5 |
NEK6
|
NIMA-related kinase 6 |
| chr20_+_19867150 | 0.39 |
ENST00000255006.6
|
RIN2
|
Ras and Rab interactor 2 |
| chr11_+_60102304 | 0.39 |
ENST00000300182.4
|
MS4A6E
|
membrane-spanning 4-domains, subfamily A, member 6E |
| chr20_-_41818373 | 0.38 |
ENST00000373187.1
ENST00000356100.2 ENST00000373184.1 ENST00000373190.1 |
PTPRT
|
protein tyrosine phosphatase, receptor type, T |
| chr1_+_154300217 | 0.38 |
ENST00000368489.3
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr16_-_30134266 | 0.38 |
ENST00000484663.1
ENST00000478356.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr1_+_154301264 | 0.37 |
ENST00000341822.2
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chrX_+_115567767 | 0.36 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr13_-_53422640 | 0.36 |
ENST00000338862.4
ENST00000377942.3 |
PCDH8
|
protocadherin 8 |
| chr6_-_39290316 | 0.35 |
ENST00000425054.2
ENST00000373227.4 ENST00000373229.5 ENST00000437525.2 |
KCNK16
|
potassium channel, subfamily K, member 16 |
| chr3_+_108321623 | 0.35 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr16_-_3149229 | 0.34 |
ENST00000572431.1
ENST00000572548.1 |
ZSCAN10
|
zinc finger and SCAN domain containing 10 |
| chr2_+_136289030 | 0.34 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr7_-_28220354 | 0.33 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr6_+_26273144 | 0.33 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr10_+_111967345 | 0.33 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr16_+_57769635 | 0.33 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr10_+_133753533 | 0.32 |
ENST00000422256.2
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr12_+_123717967 | 0.32 |
ENST00000536130.1
ENST00000546132.1 |
C12orf65
|
chromosome 12 open reading frame 65 |
| chr14_-_75179774 | 0.32 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr14_+_81421355 | 0.32 |
ENST00000541158.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr15_-_72563585 | 0.32 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr7_+_142457315 | 0.31 |
ENST00000486171.1
ENST00000311737.7 |
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr7_-_14942467 | 0.30 |
ENST00000407950.1
ENST00000444700.2 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr19_+_41313017 | 0.30 |
ENST00000595621.1
ENST00000595051.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr12_-_50222187 | 0.30 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr15_-_81282133 | 0.29 |
ENST00000261758.4
|
MESDC2
|
mesoderm development candidate 2 |
| chr10_+_73156664 | 0.29 |
ENST00000398809.4
ENST00000398842.3 ENST00000461841.3 ENST00000299366.7 |
CDH23
|
cadherin-related 23 |
| chr14_+_78227105 | 0.27 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr10_+_86184676 | 0.27 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr2_+_219646462 | 0.27 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr2_-_27545431 | 0.26 |
ENST00000233545.2
|
MPV17
|
MpV17 mitochondrial inner membrane protein |
| chr4_-_168155417 | 0.26 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr7_+_99775366 | 0.25 |
ENST00000394018.2
ENST00000416412.1 |
STAG3
|
stromal antigen 3 |
| chrX_-_13835398 | 0.24 |
ENST00000475307.1
|
GPM6B
|
glycoprotein M6B |
| chr17_+_54230819 | 0.24 |
ENST00000318698.2
ENST00000566473.2 |
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr1_-_102312517 | 0.24 |
ENST00000338858.5
|
OLFM3
|
olfactomedin 3 |
| chr5_+_169780485 | 0.23 |
ENST00000377360.4
|
KCNIP1
|
Kv channel interacting protein 1 |
| chr7_-_99774945 | 0.23 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr5_+_122110691 | 0.22 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr3_-_56717246 | 0.22 |
ENST00000355628.5
|
FAM208A
|
family with sequence similarity 208, member A |
| chr20_+_57226841 | 0.21 |
ENST00000358029.4
ENST00000361830.3 |
STX16
|
syntaxin 16 |
| chr17_-_40169659 | 0.21 |
ENST00000457167.4
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr3_+_130300585 | 0.20 |
ENST00000511332.1
|
COL6A6
|
collagen, type VI, alpha 6 |
| chr3_-_107941230 | 0.20 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr15_+_42841008 | 0.20 |
ENST00000260372.3
ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2
|
HAUS augmin-like complex, subunit 2 |
| chr10_-_49813090 | 0.20 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr11_+_94277017 | 0.20 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr10_+_79626633 | 0.20 |
ENST00000372387.1
|
AL391421.1
|
Uncharacterized protein; cDNA FLJ43696 fis, clone TBAES2007964 |
| chr11_-_5863127 | 0.19 |
ENST00000329322.5
ENST00000379946.2 |
OR52E6
|
olfactory receptor, family 52, subfamily E, member 6 |
| chr7_+_151791095 | 0.18 |
ENST00000422997.2
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr1_-_26232951 | 0.18 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr1_-_23520755 | 0.17 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr12_-_51422017 | 0.17 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr13_+_25670268 | 0.17 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr8_-_95220775 | 0.17 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr1_+_203765437 | 0.17 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chrX_+_14547632 | 0.16 |
ENST00000218075.4
|
GLRA2
|
glycine receptor, alpha 2 |
| chr15_+_42697018 | 0.16 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr16_-_30134441 | 0.16 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr15_-_42840961 | 0.16 |
ENST00000563454.1
ENST00000397130.3 ENST00000570160.1 ENST00000323443.2 |
LRRC57
|
leucine rich repeat containing 57 |
| chr3_-_127309521 | 0.16 |
ENST00000469111.1
|
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chr3_+_42201653 | 0.16 |
ENST00000341421.3
ENST00000396175.1 |
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr12_+_14518598 | 0.16 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr12_+_123717458 | 0.15 |
ENST00000253233.1
|
C12orf65
|
chromosome 12 open reading frame 65 |
| chr17_+_27055798 | 0.14 |
ENST00000268766.6
|
NEK8
|
NIMA-related kinase 8 |
| chr14_-_68159152 | 0.14 |
ENST00000554035.1
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr11_+_17756279 | 0.14 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr1_+_211433275 | 0.13 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr22_-_33968239 | 0.13 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.4 | 1.7 | GO:1905229 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.4 | 8.6 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.3 | 4.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.3 | 1.7 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.3 | 1.6 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 1.0 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.2 | 1.0 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.2 | 4.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 0.7 | GO:0052364 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.2 | 2.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.2 | 0.9 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.2 | 6.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.2 | 0.5 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.2 | 12.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.4 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 3.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 2.0 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 3.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 2.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.6 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.1 | 0.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.7 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.7 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 9.9 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 0.9 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 1.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.6 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.8 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.6 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.5 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.9 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.7 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 5.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.8 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.3 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.5 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 2.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 4.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 3.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.5 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 1.2 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 1.1 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 1.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 1.5 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 1.3 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 1.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.4 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.5 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.9 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 1.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 14.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 12.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.7 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 1.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 2.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 1.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 6.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 8.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.4 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 4.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0097134 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 6.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 5.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.6 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.4 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.2 | 1.7 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.2 | 2.0 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 3.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 12.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.7 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 2.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 1.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.4 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 2.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.9 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 2.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 1.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 4.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 3.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.2 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 1.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 8.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 4.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 8.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 6.3 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 6.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 11.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 6.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 7.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 4.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 2.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |