Project
Illumina Body Map 2
Navigation
Downloads
Results for NR1I3
Z-value: 0.50
Transcription factors associated with NR1I3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I3
|
ENSG00000143257.7 | nuclear receptor subfamily 1 group I member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I3 | hg19_v2_chr1_-_161207986_161208000 | 0.26 | 1.5e-01 | Click! |
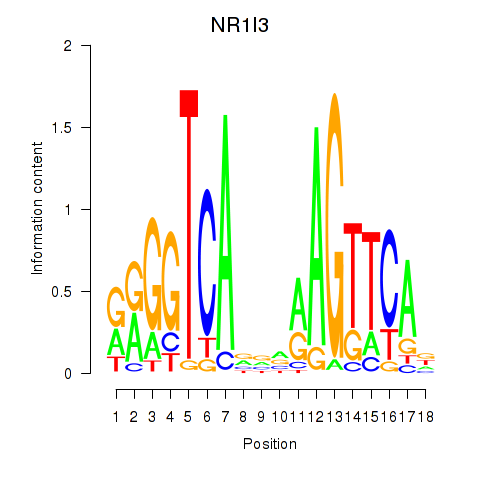
Activity profile of NR1I3 motif
Sorted Z-values of NR1I3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_57766075 | 2.54 |
ENST00000371030.2
|
ZNF831
|
zinc finger protein 831 |
| chr10_-_73848764 | 1.69 |
ENST00000317376.4
ENST00000412663.1 |
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr13_-_20806440 | 1.53 |
ENST00000400066.3
ENST00000400065.3 ENST00000356192.6 |
GJB6
|
gap junction protein, beta 6, 30kDa |
| chr22_+_42372764 | 1.39 |
ENST00000396426.3
ENST00000406029.1 |
SEPT3
|
septin 3 |
| chr7_-_31380502 | 1.30 |
ENST00000297142.3
|
NEUROD6
|
neuronal differentiation 6 |
| chr1_+_153330322 | 1.23 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr11_+_105480740 | 1.22 |
ENST00000393125.2
|
GRIA4
|
glutamate receptor, ionotropic, AMPA 4 |
| chr4_-_57524061 | 1.15 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr12_-_75601778 | 1.12 |
ENST00000550433.1
ENST00000548513.1 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr14_-_50506589 | 1.06 |
ENST00000553914.2
|
RP11-58E21.3
|
RP11-58E21.3 |
| chr1_-_235116495 | 1.05 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr8_+_11351876 | 1.01 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr5_+_156712372 | 1.01 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr5_-_39270725 | 0.85 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr22_+_39493207 | 0.83 |
ENST00000348946.4
ENST00000442487.3 ENST00000421988.2 |
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr14_+_22748980 | 0.81 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr11_+_105480770 | 0.80 |
ENST00000531986.1
|
GRIA4
|
glutamate receptor, ionotropic, AMPA 4 |
| chr5_-_176937351 | 0.78 |
ENST00000377112.4
ENST00000501403.2 ENST00000312943.6 |
DOK3
|
docking protein 3 |
| chr1_-_205290865 | 0.78 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr20_-_44007014 | 0.72 |
ENST00000372726.3
ENST00000537995.1 |
TP53TG5
|
TP53 target 5 |
| chr4_+_37245799 | 0.71 |
ENST00000309447.5
|
KIAA1239
|
KIAA1239 |
| chr12_-_54779511 | 0.71 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr6_+_32407619 | 0.70 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr11_-_83393303 | 0.68 |
ENST00000398304.1
ENST00000420775.2 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr22_+_39493268 | 0.67 |
ENST00000401756.1
|
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr6_+_108487245 | 0.60 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr19_-_49140609 | 0.59 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr12_-_53729525 | 0.51 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr16_+_58535372 | 0.51 |
ENST00000566656.1
ENST00000566618.1 |
NDRG4
|
NDRG family member 4 |
| chr13_-_95131923 | 0.51 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr5_-_176936817 | 0.51 |
ENST00000502885.1
ENST00000506493.1 |
DOK3
|
docking protein 3 |
| chr11_-_88799113 | 0.50 |
ENST00000393294.3
|
GRM5
|
glutamate receptor, metabotropic 5 |
| chr3_+_52350335 | 0.50 |
ENST00000420323.2
|
DNAH1
|
dynein, axonemal, heavy chain 1 |
| chr3_-_108476231 | 0.50 |
ENST00000295755.6
|
RETNLB
|
resistin like beta |
| chrX_-_39186610 | 0.49 |
ENST00000429281.1
ENST00000448597.1 |
RP11-265P11.2
|
RP11-265P11.2 |
| chr16_-_4465886 | 0.49 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr19_-_9968816 | 0.48 |
ENST00000590841.1
|
OLFM2
|
olfactomedin 2 |
| chr11_-_83878041 | 0.46 |
ENST00000398299.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr1_-_32210275 | 0.46 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr1_+_154378049 | 0.46 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr7_+_143079000 | 0.46 |
ENST00000392910.2
|
ZYX
|
zyxin |
| chr3_-_116163830 | 0.45 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr17_-_53499218 | 0.45 |
ENST00000571578.1
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr4_-_82136114 | 0.44 |
ENST00000395578.1
ENST00000418486.2 |
PRKG2
|
protein kinase, cGMP-dependent, type II |
| chr1_+_43803475 | 0.44 |
ENST00000372470.3
ENST00000413998.2 |
MPL
|
myeloproliferative leukemia virus oncogene |
| chr14_+_22988947 | 0.43 |
ENST00000390513.1
|
TRAJ24
|
T cell receptor alpha joining 24 |
| chrX_-_110655391 | 0.43 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr11_-_71781096 | 0.42 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr11_-_71753188 | 0.40 |
ENST00000543009.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr11_-_104480019 | 0.40 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr9_+_75766763 | 0.39 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr14_-_22005062 | 0.38 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr12_-_118810688 | 0.38 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chr7_-_56160666 | 0.38 |
ENST00000297373.2
|
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr3_-_71802760 | 0.38 |
ENST00000295612.3
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr17_-_10701241 | 0.36 |
ENST00000578763.1
|
LINC00675
|
long intergenic non-protein coding RNA 675 |
| chr11_-_61124266 | 0.36 |
ENST00000539890.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr19_-_49140692 | 0.36 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr14_-_22005018 | 0.35 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr2_-_69180083 | 0.34 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr1_-_225616515 | 0.34 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr18_+_61557781 | 0.33 |
ENST00000443281.1
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr9_-_35361262 | 0.32 |
ENST00000599954.1
|
AL160274.1
|
HCG17281; PRO0038; Uncharacterized protein |
| chr9_-_114937676 | 0.32 |
ENST00000374270.3
|
SUSD1
|
sushi domain containing 1 |
| chr1_+_145516252 | 0.32 |
ENST00000369306.3
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr1_+_154377669 | 0.31 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr17_-_53499310 | 0.31 |
ENST00000262065.3
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr18_-_19283649 | 0.31 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr8_-_143833918 | 0.30 |
ENST00000359228.3
|
LYPD2
|
LY6/PLAUR domain containing 2 |
| chr12_+_100594557 | 0.30 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr7_-_56160625 | 0.29 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr16_+_30710462 | 0.29 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr1_+_145516560 | 0.29 |
ENST00000537888.1
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr10_-_50396425 | 0.28 |
ENST00000374148.1
|
C10orf128
|
chromosome 10 open reading frame 128 |
| chr10_+_80027085 | 0.28 |
ENST00000415959.1
|
LINC00595
|
long intergenic non-protein coding RNA 595 |
| chr9_-_125148928 | 0.27 |
ENST00000600713.1
|
AL162424.1
|
Uncharacterized protein |
| chr19_-_44306590 | 0.27 |
ENST00000377950.3
|
LYPD5
|
LY6/PLAUR domain containing 5 |
| chr6_-_127780510 | 0.27 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr1_+_59250815 | 0.26 |
ENST00000544621.1
ENST00000419531.2 |
RP4-794H19.2
|
long intergenic non-protein coding RNA 1135 |
| chr2_-_69180012 | 0.26 |
ENST00000481498.1
|
GKN2
|
gastrokine 2 |
| chr4_+_70894130 | 0.25 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr18_-_19284724 | 0.25 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
| chr11_+_63137251 | 0.25 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr15_-_42343388 | 0.24 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2, group IVE |
| chr5_+_139055055 | 0.24 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr14_+_100070869 | 0.24 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr1_+_44401479 | 0.23 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr7_+_101460882 | 0.23 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr6_-_34113856 | 0.21 |
ENST00000538487.2
|
GRM4
|
glutamate receptor, metabotropic 4 |
| chr3_-_116164306 | 0.21 |
ENST00000490035.2
|
LSAMP
|
limbic system-associated membrane protein |
| chr14_-_22005197 | 0.21 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr9_+_34179003 | 0.21 |
ENST00000545103.1
ENST00000543944.1 ENST00000536252.1 ENST00000540348.1 ENST00000297661.4 ENST00000379186.4 |
UBAP1
|
ubiquitin associated protein 1 |
| chr4_-_170897045 | 0.20 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr11_-_82611448 | 0.19 |
ENST00000393399.2
ENST00000313010.3 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr1_-_21625486 | 0.19 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr10_-_50396407 | 0.19 |
ENST00000374153.2
ENST00000374151.3 |
C10orf128
|
chromosome 10 open reading frame 128 |
| chr11_+_62556596 | 0.18 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr6_+_36097992 | 0.18 |
ENST00000211287.4
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chr19_-_6604094 | 0.18 |
ENST00000597430.2
|
CD70
|
CD70 molecule |
| chr15_+_43985084 | 0.17 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr17_-_33390667 | 0.16 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr15_+_85523671 | 0.16 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr12_+_49208234 | 0.16 |
ENST00000540990.1
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr8_-_53626974 | 0.16 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chrX_+_118602363 | 0.16 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr9_+_5450503 | 0.15 |
ENST00000381573.4
ENST00000381577.3 |
CD274
|
CD274 molecule |
| chrX_+_69674943 | 0.14 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr5_+_139055021 | 0.14 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr16_+_67906919 | 0.13 |
ENST00000358933.5
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr16_+_67280799 | 0.13 |
ENST00000566345.2
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr14_+_21525981 | 0.12 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chr14_+_22977587 | 0.12 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr2_-_241075706 | 0.12 |
ENST00000607357.1
ENST00000307266.3 |
MYEOV2
|
myeloma overexpressed 2 |
| chr1_+_26036093 | 0.11 |
ENST00000374329.1
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1 |
| chr1_+_160097462 | 0.10 |
ENST00000447527.1
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr11_-_59950519 | 0.10 |
ENST00000528851.1
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr1_+_32930647 | 0.10 |
ENST00000609129.1
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chrX_-_153583257 | 0.10 |
ENST00000438732.1
|
FLNA
|
filamin A, alpha |
| chr20_-_62339315 | 0.09 |
ENST00000440854.1
ENST00000607873.1 |
ARFRP1
|
ADP-ribosylation factor related protein 1 |
| chr19_+_58919992 | 0.09 |
ENST00000306910.4
ENST00000598901.1 ENST00000593920.1 ENST00000596281.1 |
ZNF584
|
zinc finger protein 584 |
| chr12_+_57998400 | 0.08 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr9_-_80646374 | 0.08 |
ENST00000286548.4
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr20_-_62339190 | 0.07 |
ENST00000324228.2
ENST00000609142.1 |
ARFRP1
|
ADP-ribosylation factor related protein 1 |
| chr10_+_47658234 | 0.07 |
ENST00000447511.2
ENST00000537271.1 |
ANTXRL
|
anthrax toxin receptor-like |
| chr17_+_62503073 | 0.07 |
ENST00000580188.1
ENST00000581056.1 |
CEP95
|
centrosomal protein 95kDa |
| chr2_-_86790593 | 0.06 |
ENST00000263856.4
ENST00000409225.2 |
CHMP3
|
charged multivesicular body protein 3 |
| chr1_+_113217043 | 0.06 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr15_+_43885252 | 0.06 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr9_+_37486005 | 0.05 |
ENST00000377792.3
|
POLR1E
|
polymerase (RNA) I polypeptide E, 53kDa |
| chr20_+_62339381 | 0.05 |
ENST00000355969.6
ENST00000357119.4 ENST00000431125.1 ENST00000369967.3 ENST00000328969.5 |
ZGPAT
|
zinc finger, CCCH-type with G patch domain |
| chr12_+_53693812 | 0.05 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chr2_-_86790472 | 0.04 |
ENST00000409727.1
|
CHMP3
|
charged multivesicular body protein 3 |
| chr11_+_933555 | 0.04 |
ENST00000534485.1
|
AP2A2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr19_+_58920102 | 0.04 |
ENST00000599238.1
ENST00000322834.7 |
ZNF584
|
zinc finger protein 584 |
| chr1_-_1690014 | 0.03 |
ENST00000400922.2
ENST00000342348.5 |
NADK
|
NAD kinase |
| chr1_-_20755140 | 0.03 |
ENST00000418743.1
ENST00000426428.1 |
RP4-749H3.1
|
long intergenic non-protein coding RNA 1141 |
| chr14_+_53173910 | 0.02 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr6_+_96969672 | 0.02 |
ENST00000369278.4
|
UFL1
|
UFM1-specific ligase 1 |
| chr19_+_7562431 | 0.01 |
ENST00000361664.2
|
C19orf45
|
chromosome 19 open reading frame 45 |
| chr14_+_53173890 | 0.01 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 0.8 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.2 | 1.5 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.2 | 0.7 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.6 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.4 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.4 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 1.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.4 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.1 | 0.5 | GO:1902938 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 0.7 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.8 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 1.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.5 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 1.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.2 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 1.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.0 | 1.3 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.2 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 2.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.2 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.2 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 1.3 | GO:0046710 | GDP metabolic process(GO:0046710) receptor localization to synapse(GO:0097120) |
| 0.0 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.5 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 1.5 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 1.0 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.8 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.2 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 2.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 1.7 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.2 | 1.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 1.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 2.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.3 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.6 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 2.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.5 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 1.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 1.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 2.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |