Project
Illumina Body Map 2
Navigation
Downloads
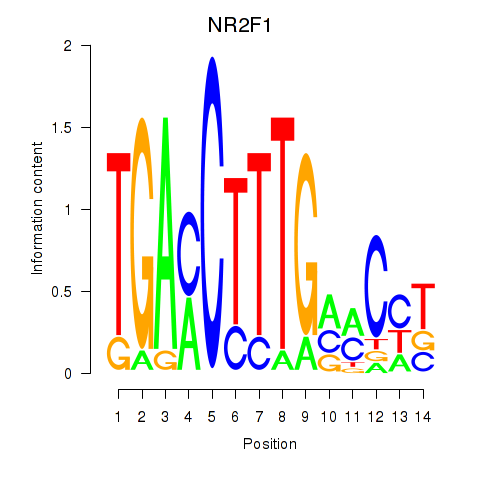
Results for NR2F1
Z-value: 1.17
Transcription factors associated with NR2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2F1
|
ENSG00000175745.7 | nuclear receptor subfamily 2 group F member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2F1 | hg19_v2_chr5_+_92919043_92919082 | 0.05 | 7.8e-01 | Click! |
Activity profile of NR2F1 motif
Sorted Z-values of NR2F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_173886491 | 7.70 |
ENST00000367698.3
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr11_-_116708302 | 5.13 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr3_-_42917363 | 4.86 |
ENST00000437102.1
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1 |
| chr16_+_56685796 | 4.75 |
ENST00000334346.2
ENST00000562399.1 |
MT1B
|
metallothionein 1B |
| chr20_+_56136136 | 4.71 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr10_-_129691195 | 4.66 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr17_+_1646130 | 4.55 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr1_+_196857144 | 4.53 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr11_-_116663127 | 4.41 |
ENST00000433069.1
ENST00000542499.1 |
APOA5
|
apolipoprotein A-V |
| chrX_+_138612889 | 4.38 |
ENST00000218099.2
ENST00000394090.2 |
F9
|
coagulation factor IX |
| chr10_-_96829246 | 4.25 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr11_-_116694009 | 4.10 |
ENST00000357780.3
|
APOA4
|
apolipoprotein A-IV |
| chr20_-_7921090 | 4.03 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr10_+_96443378 | 3.96 |
ENST00000285979.6
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr1_-_161193349 | 3.90 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr10_+_96443204 | 3.80 |
ENST00000339022.5
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr3_+_46538981 | 3.72 |
ENST00000296142.3
|
RTP3
|
receptor (chemosensory) transporter protein 3 |
| chr17_-_36105009 | 3.62 |
ENST00000560016.1
ENST00000427275.2 ENST00000561193.1 |
HNF1B
|
HNF1 homeobox B |
| chr4_+_69962185 | 3.59 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr17_+_7531281 | 3.56 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr10_-_74714533 | 3.54 |
ENST00000373032.3
|
PLA2G12B
|
phospholipase A2, group XIIB |
| chr4_+_69962212 | 3.51 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr17_+_7533439 | 3.40 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr1_+_196946680 | 3.35 |
ENST00000256785.4
|
CFHR5
|
complement factor H-related 5 |
| chr1_+_196946664 | 3.32 |
ENST00000367414.5
|
CFHR5
|
complement factor H-related 5 |
| chr7_-_99277610 | 3.30 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr6_+_30130969 | 3.09 |
ENST00000376694.4
|
TRIM15
|
tripartite motif containing 15 |
| chr10_-_54531406 | 3.09 |
ENST00000373968.3
|
MBL2
|
mannose-binding lectin (protein C) 2, soluble |
| chrX_+_106163626 | 3.00 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr1_-_11107280 | 2.94 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr1_-_169555779 | 2.90 |
ENST00000367797.3
ENST00000367796.3 |
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr3_+_113616317 | 2.84 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr19_-_51017127 | 2.79 |
ENST00000389208.4
|
ASPDH
|
aspartate dehydrogenase domain containing |
| chr14_+_103573853 | 2.71 |
ENST00000560304.1
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr22_+_25003626 | 2.70 |
ENST00000451366.1
ENST00000406383.2 ENST00000428855.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr1_-_155270770 | 2.70 |
ENST00000392414.3
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr20_-_7238861 | 2.62 |
ENST00000428954.1
|
RP11-19D2.1
|
RP11-19D2.1 |
| chr4_+_100495864 | 2.62 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr17_+_27071002 | 2.54 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr12_-_21757774 | 2.51 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr1_-_55089191 | 2.46 |
ENST00000302250.2
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151, member A |
| chr19_-_36304201 | 2.44 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr1_-_155271213 | 2.39 |
ENST00000342741.4
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr19_-_59023348 | 2.35 |
ENST00000601355.1
ENST00000263093.2 |
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr7_-_99332719 | 2.34 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr2_-_169887827 | 2.29 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr1_+_16068917 | 2.28 |
ENST00000375782.1
|
TMEM82
|
transmembrane protein 82 |
| chr2_+_228678550 | 2.25 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr19_+_5823813 | 2.24 |
ENST00000303212.2
|
NRTN
|
neurturin |
| chr10_+_96522361 | 2.21 |
ENST00000371321.3
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr10_+_101542462 | 2.19 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr12_-_14996355 | 2.17 |
ENST00000228936.4
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr17_-_34329084 | 2.15 |
ENST00000354059.4
ENST00000536149.1 |
CCL15
CCL14
|
chemokine (C-C motif) ligand 15 chemokine (C-C motif) ligand 14 |
| chr10_-_91403625 | 2.10 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr12_+_121416489 | 2.09 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr22_+_25003606 | 2.08 |
ENST00000432867.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr1_+_171060018 | 2.06 |
ENST00000367755.4
ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3
|
flavin containing monooxygenase 3 |
| chr17_-_34345002 | 2.04 |
ENST00000293280.2
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr2_-_28113217 | 2.00 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr17_-_7531121 | 1.99 |
ENST00000573566.1
ENST00000269298.5 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr1_-_169555709 | 1.98 |
ENST00000546081.1
|
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr1_-_203144941 | 1.95 |
ENST00000255416.4
|
MYBPH
|
myosin binding protein H |
| chr22_+_25003568 | 1.91 |
ENST00000447416.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr19_+_41594377 | 1.90 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr6_+_31887761 | 1.87 |
ENST00000413154.1
|
C2
|
complement component 2 |
| chr17_-_34344991 | 1.85 |
ENST00000591423.1
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr2_+_220363579 | 1.77 |
ENST00000313597.5
ENST00000373917.3 ENST00000358215.3 ENST00000373908.1 ENST00000455657.1 ENST00000435316.1 ENST00000341142.3 |
GMPPA
|
GDP-mannose pyrophosphorylase A |
| chr2_+_44502630 | 1.76 |
ENST00000410056.3
ENST00000409741.1 ENST00000409229.3 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr1_-_197036364 | 1.70 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr14_+_21492331 | 1.70 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr4_-_40477766 | 1.66 |
ENST00000507180.1
|
RBM47
|
RNA binding motif protein 47 |
| chr6_+_33172407 | 1.64 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr11_+_2920951 | 1.62 |
ENST00000347936.2
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr19_+_7660716 | 1.62 |
ENST00000160298.4
ENST00000446248.2 |
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr19_+_6464243 | 1.61 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr1_-_48227290 | 1.59 |
ENST00000594280.1
|
FLJ00388
|
FLJ00388 |
| chr12_-_7244469 | 1.57 |
ENST00000538050.1
ENST00000536053.2 |
C1R
|
complement component 1, r subcomponent |
| chr11_-_72493574 | 1.56 |
ENST00000536290.1
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr7_-_15601595 | 1.53 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr7_+_97840739 | 1.50 |
ENST00000609256.1
|
BHLHA15
|
basic helix-loop-helix family, member a15 |
| chr19_+_6464502 | 1.50 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr11_+_278365 | 1.49 |
ENST00000534750.1
|
NLRP6
|
NLR family, pyrin domain containing 6 |
| chr1_+_158149737 | 1.44 |
ENST00000368171.3
|
CD1D
|
CD1d molecule |
| chr15_+_58724184 | 1.44 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr19_-_16045619 | 1.39 |
ENST00000402119.4
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr19_-_16045665 | 1.36 |
ENST00000248041.8
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr2_-_165698521 | 1.35 |
ENST00000409184.3
ENST00000392717.2 ENST00000456693.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr8_-_144815966 | 1.34 |
ENST00000388913.3
|
FAM83H
|
family with sequence similarity 83, member H |
| chrX_+_38211777 | 1.30 |
ENST00000039007.4
|
OTC
|
ornithine carbamoyltransferase |
| chr1_+_53793885 | 1.28 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr12_+_121416340 | 1.26 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr3_-_53878644 | 1.25 |
ENST00000481668.1
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr2_+_219646462 | 1.24 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr3_-_126236588 | 1.23 |
ENST00000383579.3
|
UROC1
|
urocanate hydratase 1 |
| chrX_+_66764375 | 1.19 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr9_+_6716478 | 1.18 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chrX_-_114253536 | 1.18 |
ENST00000371936.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr2_-_165698662 | 1.17 |
ENST00000194871.6
ENST00000445474.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr17_-_47786375 | 1.13 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr12_-_7125770 | 1.12 |
ENST00000261407.4
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3 |
| chr3_-_126236605 | 1.11 |
ENST00000290868.2
|
UROC1
|
urocanate hydratase 1 |
| chr1_+_101185290 | 1.11 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr11_+_18417813 | 1.10 |
ENST00000540430.1
ENST00000379412.5 |
LDHA
|
lactate dehydrogenase A |
| chr8_-_128231299 | 1.08 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr1_+_176432298 | 1.07 |
ENST00000367661.3
ENST00000367662.3 |
PAPPA2
|
pappalysin 2 |
| chr13_+_113777105 | 1.04 |
ENST00000409306.1
ENST00000375551.3 ENST00000375559.3 |
F10
|
coagulation factor X |
| chr19_+_49713991 | 1.04 |
ENST00000597316.1
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr12_+_121416437 | 1.03 |
ENST00000402929.1
ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A
|
HNF1 homeobox A |
| chr2_-_241836244 | 1.01 |
ENST00000454476.2
|
C2orf54
|
chromosome 2 open reading frame 54 |
| chr19_-_58892389 | 1.01 |
ENST00000427624.2
ENST00000597582.1 |
ZNF837
|
zinc finger protein 837 |
| chr7_+_65552756 | 1.00 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr2_+_197577841 | 0.99 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr2_-_241836298 | 0.99 |
ENST00000414499.1
|
C2orf54
|
chromosome 2 open reading frame 54 |
| chr5_-_42812143 | 0.97 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr4_-_109683691 | 0.96 |
ENST00000512320.1
ENST00000510723.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr12_+_132413765 | 0.95 |
ENST00000376649.3
ENST00000322060.5 |
PUS1
|
pseudouridylate synthase 1 |
| chr5_-_95158644 | 0.94 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr16_+_2076869 | 0.93 |
ENST00000424542.2
ENST00000432365.2 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr21_-_45682099 | 0.91 |
ENST00000270172.3
ENST00000418993.1 |
DNMT3L
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr2_-_70780770 | 0.90 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr12_+_132413739 | 0.90 |
ENST00000443358.2
|
PUS1
|
pseudouridylate synthase 1 |
| chr16_-_31439735 | 0.90 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr2_-_198364552 | 0.89 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr4_+_159593418 | 0.89 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr6_+_143381594 | 0.89 |
ENST00000367601.4
|
AIG1
|
androgen-induced 1 |
| chr10_+_114133773 | 0.88 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr2_+_219283815 | 0.88 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr17_+_4853442 | 0.88 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr11_+_63137251 | 0.87 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr7_+_192969 | 0.87 |
ENST00000313766.5
|
FAM20C
|
family with sequence similarity 20, member C |
| chr5_-_42811986 | 0.87 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chrX_-_128977364 | 0.86 |
ENST00000371064.3
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr16_+_83932684 | 0.84 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr16_+_21244986 | 0.83 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr12_-_56727676 | 0.82 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr14_-_23284703 | 0.81 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr9_-_27005686 | 0.81 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr2_+_138721850 | 0.81 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr16_-_30997533 | 0.80 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr2_-_198364581 | 0.80 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr2_+_198365095 | 0.77 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr14_-_23285069 | 0.77 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr12_-_56727487 | 0.77 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr14_-_23285011 | 0.74 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr11_-_31014214 | 0.74 |
ENST00000406071.2
ENST00000339794.5 |
DCDC1
|
doublecortin domain containing 1 |
| chr12_-_56367101 | 0.73 |
ENST00000549233.2
|
PMEL
|
premelanosome protein |
| chr1_+_33207381 | 0.72 |
ENST00000401073.2
|
KIAA1522
|
KIAA1522 |
| chr17_-_2614927 | 0.71 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr14_+_24590560 | 0.71 |
ENST00000558325.1
|
RP11-468E2.6
|
RP11-468E2.6 |
| chr6_+_43737939 | 0.70 |
ENST00000372067.3
|
VEGFA
|
vascular endothelial growth factor A |
| chr13_+_100741269 | 0.70 |
ENST00000376286.4
ENST00000376279.3 ENST00000376285.1 |
PCCA
|
propionyl CoA carboxylase, alpha polypeptide |
| chr6_+_7108210 | 0.68 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr5_+_118965244 | 0.68 |
ENST00000515256.1
ENST00000509264.1 |
FAM170A
|
family with sequence similarity 170, member A |
| chr19_+_10362882 | 0.67 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr1_-_43751276 | 0.67 |
ENST00000423420.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr7_-_100844193 | 0.66 |
ENST00000440203.2
ENST00000379423.3 ENST00000223114.4 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr19_-_38806390 | 0.66 |
ENST00000589247.1
ENST00000329420.8 ENST00000591784.1 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr13_+_76413852 | 0.65 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr19_-_38806540 | 0.64 |
ENST00000592694.1
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr17_-_7307358 | 0.64 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr4_-_141075330 | 0.63 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr17_-_80017856 | 0.63 |
ENST00000577574.1
|
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr12_+_132413798 | 0.62 |
ENST00000440818.2
ENST00000542167.2 ENST00000538037.1 ENST00000456665.2 |
PUS1
|
pseudouridylate synthase 1 |
| chr8_-_70016408 | 0.61 |
ENST00000518540.1
|
RP11-600K15.1
|
RP11-600K15.1 |
| chr12_+_6494285 | 0.61 |
ENST00000541102.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr11_-_124543725 | 0.60 |
ENST00000545756.1
ENST00000263593.3 |
SIAE
|
sialic acid acetylesterase |
| chr11_-_26743546 | 0.60 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr19_-_39303576 | 0.59 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr7_+_107301065 | 0.58 |
ENST00000265715.3
|
SLC26A4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr11_+_12108410 | 0.58 |
ENST00000527997.1
|
RP13-631K18.5
|
RP13-631K18.5 |
| chr2_+_44502597 | 0.57 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr1_+_28696111 | 0.57 |
ENST00000373839.3
|
PHACTR4
|
phosphatase and actin regulator 4 |
| chr6_+_30131318 | 0.55 |
ENST00000376688.1
|
TRIM15
|
tripartite motif containing 15 |
| chr3_+_113465866 | 0.55 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr8_-_80942467 | 0.54 |
ENST00000518271.1
ENST00000276585.4 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr14_-_23284675 | 0.54 |
ENST00000555959.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr2_-_73460334 | 0.54 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr15_+_90728145 | 0.53 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr3_-_171178157 | 0.53 |
ENST00000465393.1
ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr3_-_139195350 | 0.53 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr9_-_95896550 | 0.52 |
ENST00000375446.4
|
NINJ1
|
ninjurin 1 |
| chr9_-_98079154 | 0.51 |
ENST00000433829.1
|
FANCC
|
Fanconi anemia, complementation group C |
| chr19_-_38806560 | 0.51 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr10_+_81107271 | 0.51 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr11_-_1776176 | 0.51 |
ENST00000429746.1
|
CTSD
|
cathepsin D |
| chr9_-_127358087 | 0.51 |
ENST00000475178.1
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr19_+_10197463 | 0.51 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr2_-_85581701 | 0.49 |
ENST00000295802.4
|
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr8_-_37594944 | 0.49 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr20_-_62601218 | 0.49 |
ENST00000369888.1
|
ZNF512B
|
zinc finger protein 512B |
| chr2_+_198365122 | 0.48 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr22_-_47882857 | 0.48 |
ENST00000405369.3
|
LL22NC03-75H12.2
|
Novel protein; Uncharacterized protein |
| chr6_+_32146131 | 0.48 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr7_-_105926058 | 0.47 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr17_-_2615031 | 0.46 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr3_-_141747459 | 0.46 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr16_+_4475806 | 0.46 |
ENST00000262375.6
ENST00000355296.4 ENST00000431375.2 ENST00000574895.1 |
DNAJA3
|
DnaJ (Hsp40) homolog, subfamily A, member 3 |
| chr10_-_74114714 | 0.46 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr22_-_24641027 | 0.46 |
ENST00000398292.3
ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5
|
gamma-glutamyltransferase 5 |
| chr2_-_85581623 | 0.45 |
ENST00000449375.1
ENST00000409984.2 ENST00000457495.2 ENST00000263854.6 |
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 2.0 | 8.0 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 1.6 | 4.7 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 1.4 | 9.9 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 1.2 | 8.5 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 1.2 | 3.6 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.9 | 2.8 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.8 | 2.5 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.8 | 4.9 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.8 | 5.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.8 | 2.3 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.7 | 4.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.7 | 2.7 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.7 | 2.0 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.7 | 4.6 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.6 | 6.7 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.6 | 1.7 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.5 | 5.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.5 | 4.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.5 | 11.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.5 | 5.2 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.4 | 6.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.4 | 1.3 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.4 | 2.0 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.4 | 1.2 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.4 | 1.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.4 | 1.1 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.3 | 2.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 2.3 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.3 | 1.3 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.3 | 2.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.3 | 2.6 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.3 | 0.9 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.3 | 7.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.3 | 7.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.3 | 1.1 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.3 | 3.1 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.3 | 1.0 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.2 | 0.7 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.2 | 0.8 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.2 | 1.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.2 | 1.6 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 1.6 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.2 | 0.6 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 0.9 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.5 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.2 | 2.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 1.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 2.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 1.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 1.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 4.8 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 2.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.8 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.8 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 1.4 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 1.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.4 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.8 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 1.2 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 0.7 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 2.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.9 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 1.7 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.1 | 5.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 3.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.9 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 1.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 3.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 5.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.2 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 2.2 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.1 | 0.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 3.5 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 1.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.2 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.6 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 2.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.1 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.1 | 1.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.7 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.6 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 1.1 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.2 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 6.0 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.7 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.9 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 1.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.9 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 1.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.5 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.5 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 3.1 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 2.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.2 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 1.6 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 2.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.8 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 1.1 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.6 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.4 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.5 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.9 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.7 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.3 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.3 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.7 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 1.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.5 | 6.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.5 | 12.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 1.6 | GO:0031251 | PAN complex(GO:0031251) |
| 0.4 | 1.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.3 | 3.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.3 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 2.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.3 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 1.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) plasma membrane respiratory chain(GO:0070470) |
| 0.1 | 7.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 4.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 1.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 2.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 8.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.5 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.6 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 1.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 13.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 39.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 3.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 7.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0001669 | acrosomal vesicle(GO:0001669) sperm part(GO:0097223) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.6 | 4.7 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 1.3 | 4.0 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 1.1 | 8.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.8 | 4.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.8 | 5.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.8 | 2.5 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.8 | 8.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.8 | 2.3 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.8 | 2.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.7 | 2.9 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.7 | 15.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.7 | 2.0 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.6 | 2.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.6 | 3.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.5 | 1.6 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.5 | 3.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.4 | 7.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 1.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.3 | 8.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 2.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 2.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.3 | 0.9 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.3 | 1.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.3 | 1.7 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.3 | 1.6 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.2 | 1.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 2.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 2.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 2.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 1.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 0.4 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.2 | 2.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 0.5 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.2 | 1.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.4 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 3.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 7.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.8 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 2.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 1.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 2.5 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 2.8 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.1 | 1.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.7 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.4 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.6 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 12.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.6 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 2.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 3.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.7 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 1.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 3.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 4.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 2.0 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 1.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 2.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 4.5 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 1.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 1.0 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 6.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 3.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.3 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 22.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 8.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 8.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 8.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 13.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 2.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 21.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.7 | 15.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.6 | 17.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 8.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 6.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.4 | 4.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 4.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 7.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 9.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 3.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 7.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 6.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 5.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 5.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 5.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 2.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 5.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |