Project
Illumina Body Map 2
Navigation
Downloads
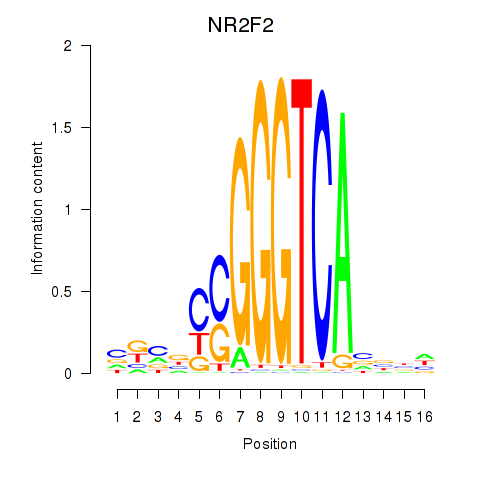
Results for NR2F2
Z-value: 1.43
Transcription factors associated with NR2F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2F2
|
ENSG00000185551.8 | nuclear receptor subfamily 2 group F member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2F2 | hg19_v2_chr15_+_96875657_96875794 | 0.16 | 3.7e-01 | Click! |
Activity profile of NR2F2 motif
Sorted Z-values of NR2F2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_137365402 | 3.17 |
ENST00000541547.1
|
IL20RA
|
interleukin 20 receptor, alpha |
| chr21_-_43187231 | 3.10 |
ENST00000332512.3
ENST00000352483.2 |
RIPK4
|
receptor-interacting serine-threonine kinase 4 |
| chr19_-_4517613 | 2.62 |
ENST00000301286.3
|
PLIN4
|
perilipin 4 |
| chr1_-_153588765 | 2.53 |
ENST00000368701.1
ENST00000344616.2 |
S100A14
|
S100 calcium binding protein A14 |
| chr11_+_1093318 | 2.50 |
ENST00000333592.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr15_+_33010175 | 2.32 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr21_-_42880075 | 2.25 |
ENST00000332149.5
|
TMPRSS2
|
transmembrane protease, serine 2 |
| chr21_-_42879909 | 2.19 |
ENST00000458356.1
ENST00000398585.3 ENST00000424093.1 |
TMPRSS2
|
transmembrane protease, serine 2 |
| chr4_-_77819002 | 1.95 |
ENST00000334306.2
|
SOWAHB
|
sosondowah ankyrin repeat domain family member B |
| chr6_-_137366096 | 1.90 |
ENST00000316649.5
ENST00000367746.3 |
IL20RA
|
interleukin 20 receptor, alpha |
| chr6_+_19837592 | 1.75 |
ENST00000378700.3
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr2_+_169659121 | 1.70 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr20_-_52687059 | 1.63 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr6_-_137366163 | 1.62 |
ENST00000367748.1
|
IL20RA
|
interleukin 20 receptor, alpha |
| chrX_-_21776281 | 1.59 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr2_+_169658928 | 1.57 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr7_+_116139744 | 1.54 |
ENST00000343213.2
|
CAV2
|
caveolin 2 |
| chr2_-_193059634 | 1.52 |
ENST00000392314.1
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr3_-_142608001 | 1.52 |
ENST00000295992.3
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr3_-_142607740 | 1.47 |
ENST00000485766.1
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr15_+_51973680 | 1.47 |
ENST00000542355.2
|
SCG3
|
secretogranin III |
| chr18_-_6929797 | 1.47 |
ENST00000581725.1
ENST00000583316.1 |
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr7_+_79764104 | 1.46 |
ENST00000351004.3
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr7_+_116139821 | 1.45 |
ENST00000393480.2
|
CAV2
|
caveolin 2 |
| chr4_-_154710210 | 1.45 |
ENST00000274063.4
|
SFRP2
|
secreted frizzled-related protein 2 |
| chr7_+_97361388 | 1.43 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chr19_-_45826125 | 1.43 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr11_+_18287721 | 1.42 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr1_-_21978312 | 1.41 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr11_+_1944054 | 1.41 |
ENST00000397301.1
ENST00000397304.2 ENST00000446240.1 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr2_+_102928009 | 1.40 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr12_+_52626898 | 1.40 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr7_-_73038822 | 1.35 |
ENST00000414749.2
ENST00000429400.2 ENST00000434326.1 |
MLXIPL
|
MLX interacting protein-like |
| chr17_-_42031300 | 1.32 |
ENST00000592796.1
|
PYY
|
peptide YY |
| chr20_+_43343886 | 1.32 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr7_-_73038867 | 1.31 |
ENST00000313375.3
ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein-like |
| chr2_+_189157536 | 1.31 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr3_-_196695692 | 1.31 |
ENST00000412723.1
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis, class Z |
| chr7_+_97361218 | 1.25 |
ENST00000319273.5
|
TAC1
|
tachykinin, precursor 1 |
| chr1_+_43996518 | 1.24 |
ENST00000359947.4
ENST00000438120.1 |
PTPRF
|
protein tyrosine phosphatase, receptor type, F |
| chr2_-_241737128 | 1.23 |
ENST00000404283.3
|
KIF1A
|
kinesin family member 1A |
| chr1_-_95007193 | 1.23 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr2_+_189157498 | 1.22 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr19_-_43702231 | 1.21 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr15_+_51973708 | 1.21 |
ENST00000558709.1
|
SCG3
|
secretogranin III |
| chr10_-_97321112 | 1.20 |
ENST00000607232.1
ENST00000371227.4 ENST00000371249.2 ENST00000371247.2 ENST00000371246.2 ENST00000393949.1 ENST00000353505.5 ENST00000347291.4 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr3_+_239652 | 1.19 |
ENST00000435603.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr17_+_41006095 | 1.17 |
ENST00000591562.1
ENST00000588033.1 |
AOC3
|
amine oxidase, copper containing 3 |
| chr10_-_104179682 | 1.17 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr7_+_95401877 | 1.17 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr8_-_57359131 | 1.17 |
ENST00000518974.1
ENST00000523051.1 ENST00000518770.1 ENST00000451791.2 |
PENK
|
proenkephalin |
| chr19_-_6720686 | 1.16 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr8_+_67344710 | 1.15 |
ENST00000379385.4
ENST00000396623.3 ENST00000415254.1 |
ADHFE1
|
alcohol dehydrogenase, iron containing, 1 |
| chr5_+_63461642 | 1.11 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr5_+_126626498 | 1.11 |
ENST00000503335.2
ENST00000508365.1 ENST00000418761.2 ENST00000274473.6 |
MEGF10
|
multiple EGF-like-domains 10 |
| chr10_-_97321165 | 1.10 |
ENST00000306402.6
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr1_+_215256467 | 1.09 |
ENST00000391894.2
ENST00000444842.2 |
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr2_-_151344172 | 1.08 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chrX_+_99899180 | 1.08 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr3_-_50340996 | 1.08 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr12_-_16035195 | 1.07 |
ENST00000535752.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr22_-_44258280 | 1.06 |
ENST00000540422.1
|
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr9_+_124030338 | 1.05 |
ENST00000449773.1
ENST00000432226.1 ENST00000436847.1 ENST00000394353.2 ENST00000449733.1 ENST00000412819.1 ENST00000341272.2 ENST00000373808.2 |
GSN
|
gelsolin |
| chr3_-_164914640 | 1.05 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr5_-_131562935 | 1.05 |
ENST00000379104.2
ENST00000379100.2 ENST00000428369.1 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr5_-_131563501 | 1.05 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr14_+_69726656 | 1.05 |
ENST00000337827.4
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr22_+_44319648 | 1.02 |
ENST00000423180.2
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr11_-_70963538 | 1.02 |
ENST00000413503.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr17_-_37382105 | 1.01 |
ENST00000333461.5
|
STAC2
|
SH3 and cysteine rich domain 2 |
| chr6_+_54711533 | 1.01 |
ENST00000306858.7
|
FAM83B
|
family with sequence similarity 83, member B |
| chr11_-_9113137 | 1.00 |
ENST00000520467.1
ENST00000309263.3 ENST00000457346.2 |
SCUBE2
|
signal peptide, CUB domain, EGF-like 2 |
| chr7_+_107301447 | 0.99 |
ENST00000440056.1
|
SLC26A4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr2_+_95940220 | 0.99 |
ENST00000542147.1
|
PROM2
|
prominin 2 |
| chr1_+_42619070 | 0.99 |
ENST00000372581.1
|
GUCA2B
|
guanylate cyclase activator 2B (uroguanylin) |
| chrX_+_2670066 | 0.98 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chr15_-_37393406 | 0.97 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr12_-_85306562 | 0.96 |
ENST00000551612.1
ENST00000450363.3 ENST00000552192.1 |
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr19_-_7990991 | 0.96 |
ENST00000318978.4
|
CTXN1
|
cortexin 1 |
| chr3_+_145782358 | 0.95 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr5_-_174871136 | 0.95 |
ENST00000393752.2
|
DRD1
|
dopamine receptor D1 |
| chr11_+_18287801 | 0.95 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr10_-_71892555 | 0.94 |
ENST00000307864.1
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr1_+_215740709 | 0.93 |
ENST00000259154.4
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr16_+_55522536 | 0.93 |
ENST00000570283.1
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr5_+_59783540 | 0.93 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr6_-_105627735 | 0.91 |
ENST00000254765.3
|
POPDC3
|
popeye domain containing 3 |
| chr12_-_85306594 | 0.91 |
ENST00000266682.5
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr21_+_45553484 | 0.91 |
ENST00000291577.6
ENST00000427803.2 |
C21orf33
|
chromosome 21 open reading frame 33 |
| chr3_-_196695733 | 0.90 |
ENST00000443835.1
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis, class Z |
| chr22_-_44258360 | 0.90 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chrX_-_43741594 | 0.90 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr18_-_30716038 | 0.90 |
ENST00000581852.1
|
CCDC178
|
coiled-coil domain containing 178 |
| chr1_-_33366931 | 0.90 |
ENST00000373463.3
ENST00000329151.5 |
TMEM54
|
transmembrane protein 54 |
| chr19_-_11529980 | 0.90 |
ENST00000567080.1
ENST00000393423.3 |
RGL3
|
ral guanine nucleotide dissociation stimulator-like 3 |
| chr14_+_69726864 | 0.89 |
ENST00000448469.3
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chrX_+_100333709 | 0.89 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chr2_+_201450591 | 0.88 |
ENST00000374700.2
|
AOX1
|
aldehyde oxidase 1 |
| chr11_-_35441597 | 0.88 |
ENST00000395753.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr19_+_13122980 | 0.88 |
ENST00000590027.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr5_+_131593364 | 0.87 |
ENST00000253754.3
ENST00000379018.3 |
PDLIM4
|
PDZ and LIM domain 4 |
| chr4_-_186697044 | 0.86 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_264024 | 0.86 |
ENST00000403712.2
ENST00000356150.5 ENST00000405430.1 |
SH3YL1
|
SH3 and SYLF domain containing 1 |
| chr3_+_157261035 | 0.86 |
ENST00000312275.5
|
C3orf55
|
chromosome 3 open reading frame 55 |
| chr11_-_35441524 | 0.85 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr9_+_116111794 | 0.85 |
ENST00000374183.4
|
BSPRY
|
B-box and SPRY domain containing |
| chr14_+_69726968 | 0.85 |
ENST00000553669.1
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr14_-_103987679 | 0.85 |
ENST00000553610.1
|
CKB
|
creatine kinase, brain |
| chr7_+_30960915 | 0.84 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr16_+_69458537 | 0.84 |
ENST00000515314.1
ENST00000561792.1 ENST00000568237.1 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr19_-_11530012 | 0.84 |
ENST00000380456.3
|
RGL3
|
ral guanine nucleotide dissociation stimulator-like 3 |
| chr3_+_238969 | 0.83 |
ENST00000421198.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr1_+_33207381 | 0.83 |
ENST00000401073.2
|
KIAA1522
|
KIAA1522 |
| chr2_+_192110199 | 0.82 |
ENST00000304164.4
|
MYO1B
|
myosin IB |
| chr12_+_112191694 | 0.82 |
ENST00000546840.2
|
RP11-162P23.2
|
RP11-162P23.2 |
| chr2_-_128408105 | 0.82 |
ENST00000409254.1
|
LIMS2
|
LIM and senescent cell antigen-like domains 2 |
| chr17_+_8214130 | 0.82 |
ENST00000579439.1
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr19_+_12862604 | 0.81 |
ENST00000553030.1
|
BEST2
|
bestrophin 2 |
| chr3_+_45067659 | 0.81 |
ENST00000296130.4
|
CLEC3B
|
C-type lectin domain family 3, member B |
| chr9_+_96051469 | 0.80 |
ENST00000453718.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr10_+_102729249 | 0.80 |
ENST00000519649.1
ENST00000518124.1 |
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr21_+_47518011 | 0.80 |
ENST00000300527.4
ENST00000357838.4 ENST00000310645.5 |
COL6A2
|
collagen, type VI, alpha 2 |
| chr17_+_41005283 | 0.79 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chr10_-_104178857 | 0.79 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr11_+_1855645 | 0.79 |
ENST00000381968.3
ENST00000381978.3 |
SYT8
|
synaptotagmin VIII |
| chr11_+_705193 | 0.78 |
ENST00000527199.1
|
EPS8L2
|
EPS8-like 2 |
| chr11_-_18270182 | 0.78 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr7_+_128828713 | 0.78 |
ENST00000249373.3
|
SMO
|
smoothened, frizzled family receptor |
| chr16_+_69458428 | 0.77 |
ENST00000512062.1
ENST00000307892.8 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr2_-_86790472 | 0.77 |
ENST00000409727.1
|
CHMP3
|
charged multivesicular body protein 3 |
| chr1_-_23694794 | 0.76 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr17_+_80693427 | 0.76 |
ENST00000300784.7
|
FN3K
|
fructosamine 3 kinase |
| chr3_-_126076264 | 0.75 |
ENST00000296233.3
|
KLF15
|
Kruppel-like factor 15 |
| chr18_+_12658209 | 0.75 |
ENST00000400512.1
|
AP005482.1
|
Uncharacterized protein |
| chr2_+_11052054 | 0.75 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr1_+_179851823 | 0.75 |
ENST00000435319.4
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr5_-_178772424 | 0.75 |
ENST00000251582.7
ENST00000274609.5 |
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr1_+_39547082 | 0.74 |
ENST00000602421.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr7_-_45960850 | 0.74 |
ENST00000381083.4
ENST00000381086.5 ENST00000275521.6 |
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr16_+_67198683 | 0.73 |
ENST00000517685.1
ENST00000521374.1 ENST00000584272.1 |
HSF4
|
heat shock transcription factor 4 |
| chr17_-_18266818 | 0.73 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr2_-_10220538 | 0.72 |
ENST00000381813.4
|
CYS1
|
cystin 1 |
| chr11_-_69294647 | 0.72 |
ENST00000542064.1
|
AP000439.3
|
AP000439.3 |
| chr3_+_238456 | 0.72 |
ENST00000427688.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr7_+_1126461 | 0.72 |
ENST00000297469.3
|
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr6_-_46459099 | 0.72 |
ENST00000371374.1
|
RCAN2
|
regulator of calcineurin 2 |
| chr20_+_33589048 | 0.71 |
ENST00000446156.1
ENST00000453028.1 ENST00000435272.1 ENST00000433934.2 ENST00000456649.1 |
MYH7B
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr17_-_53809473 | 0.71 |
ENST00000575734.1
|
TMEM100
|
transmembrane protein 100 |
| chr14_+_100204027 | 0.70 |
ENST00000554479.1
ENST00000555145.1 |
EML1
|
echinoderm microtubule associated protein like 1 |
| chr4_+_159593418 | 0.70 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr2_+_26785409 | 0.70 |
ENST00000329615.3
ENST00000409392.1 |
C2orf70
|
chromosome 2 open reading frame 70 |
| chr3_+_183993797 | 0.69 |
ENST00000359140.4
ENST00000404464.3 ENST00000357474.5 |
ECE2
|
endothelin converting enzyme 2 |
| chr2_+_171627597 | 0.69 |
ENST00000429172.1
ENST00000426475.1 |
AC007405.6
|
AC007405.6 |
| chr7_+_94536898 | 0.69 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr3_-_170303845 | 0.69 |
ENST00000231706.5
|
SLC7A14
|
solute carrier family 7, member 14 |
| chr1_-_229406746 | 0.69 |
ENST00000429227.1
ENST00000436334.1 |
RP5-1061H20.4
|
RP5-1061H20.4 |
| chr21_+_45553535 | 0.68 |
ENST00000348499.5
ENST00000389690.3 ENST00000449622.1 |
C21orf33
|
chromosome 21 open reading frame 33 |
| chr17_-_30186328 | 0.68 |
ENST00000302362.6
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr3_+_13590619 | 0.68 |
ENST00000404922.3
|
FBLN2
|
fibulin 2 |
| chrX_+_152240819 | 0.68 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr11_-_35440796 | 0.68 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr1_-_156647189 | 0.68 |
ENST00000368223.3
|
NES
|
nestin |
| chr1_+_179851893 | 0.68 |
ENST00000531630.2
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr6_+_147830063 | 0.68 |
ENST00000367474.1
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chrX_-_152245978 | 0.68 |
ENST00000538162.2
|
PNMA6D
|
paraneoplastic Ma antigen family member 6D (pseudogene) |
| chr8_-_57358432 | 0.67 |
ENST00000517415.1
ENST00000314922.3 |
PENK
|
proenkephalin |
| chr9_-_139581848 | 0.67 |
ENST00000538402.1
ENST00000371694.3 |
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr6_-_127664475 | 0.67 |
ENST00000474289.2
ENST00000534442.1 ENST00000368289.2 ENST00000525745.1 ENST00000430841.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr1_+_110254850 | 0.67 |
ENST00000369812.5
ENST00000256593.3 ENST00000369813.1 |
GSTM5
|
glutathione S-transferase mu 5 |
| chr1_+_113933581 | 0.67 |
ENST00000307546.9
ENST00000369615.1 ENST00000369611.4 |
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr18_-_712618 | 0.67 |
ENST00000583771.1
ENST00000383578.3 ENST00000251101.7 |
ENOSF1
|
enolase superfamily member 1 |
| chr9_-_139581875 | 0.67 |
ENST00000371696.2
|
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr6_+_7541845 | 0.67 |
ENST00000418664.2
|
DSP
|
desmoplakin |
| chr17_-_39928106 | 0.67 |
ENST00000540235.1
|
JUP
|
junction plakoglobin |
| chr11_+_60691924 | 0.66 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr1_+_38273818 | 0.66 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr12_-_96184913 | 0.66 |
ENST00000538383.1
|
NTN4
|
netrin 4 |
| chr21_-_43771226 | 0.65 |
ENST00000291526.4
|
TFF2
|
trefoil factor 2 |
| chr20_-_3154162 | 0.65 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr17_+_7184986 | 0.65 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr1_+_179851999 | 0.65 |
ENST00000527391.1
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr19_-_17799135 | 0.65 |
ENST00000552293.1
ENST00000551649.1 ENST00000550896.1 |
UNC13A
|
unc-13 homolog A (C. elegans) |
| chr16_+_71660052 | 0.65 |
ENST00000567566.1
|
MARVELD3
|
MARVEL domain containing 3 |
| chr19_+_12862486 | 0.65 |
ENST00000549706.1
|
BEST2
|
bestrophin 2 |
| chr8_+_21899898 | 0.63 |
ENST00000518533.1
ENST00000359441.3 |
FGF17
|
fibroblast growth factor 17 |
| chr8_-_121457332 | 0.63 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chrX_-_152343394 | 0.63 |
ENST00000370261.1
|
PNMA6B
|
paraneoplastic Ma antigen family member 6B (pseudogene) |
| chr12_-_125348329 | 0.63 |
ENST00000546215.1
ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1
|
scavenger receptor class B, member 1 |
| chr17_+_12859080 | 0.63 |
ENST00000583608.1
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr21_+_47531328 | 0.63 |
ENST00000409416.1
ENST00000397763.1 |
COL6A2
|
collagen, type VI, alpha 2 |
| chr16_+_66400533 | 0.63 |
ENST00000341529.3
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr16_-_2908155 | 0.62 |
ENST00000571228.1
ENST00000161006.3 |
PRSS22
|
protease, serine, 22 |
| chrX_+_101975643 | 0.62 |
ENST00000361229.4
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9 |
| chrX_-_99986494 | 0.62 |
ENST00000372989.1
ENST00000455616.1 ENST00000454200.2 ENST00000276141.6 |
SYTL4
|
synaptotagmin-like 4 |
| chr7_-_107642348 | 0.62 |
ENST00000393561.1
|
LAMB1
|
laminin, beta 1 |
| chr8_-_99129384 | 0.62 |
ENST00000521560.1
ENST00000254878.3 |
HRSP12
|
heat-responsive protein 12 |
| chr2_+_95940186 | 0.62 |
ENST00000403131.2
ENST00000317668.4 ENST00000317620.9 |
PROM2
|
prominin 2 |
| chr8_-_99129338 | 0.62 |
ENST00000520507.1
|
HRSP12
|
heat-responsive protein 12 |
| chr1_+_87595433 | 0.61 |
ENST00000469312.2
ENST00000490006.2 |
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr7_-_143991230 | 0.61 |
ENST00000543357.1
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2F2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.8 | 2.3 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.7 | 2.0 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.5 | 1.5 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.5 | 1.4 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.5 | 1.4 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.4 | 1.7 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.4 | 1.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.4 | 1.6 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.4 | 1.2 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.4 | 1.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.4 | 3.0 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.4 | 1.8 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.4 | 1.8 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.3 | 4.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.3 | 0.6 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.3 | 0.8 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.3 | 1.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.3 | 1.0 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.2 | 0.7 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.2 | 2.7 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.2 | 0.9 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.2 | 2.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.2 | 1.1 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 | 1.1 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.2 | 0.6 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.2 | 0.6 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.2 | 1.4 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 0.2 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.2 | 0.6 | GO:0002125 | maternal aggressive behavior(GO:0002125) positive regulation of female receptivity(GO:0045925) |
| 0.2 | 1.0 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.2 | 0.9 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.2 | 2.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 1.9 | GO:0015820 | leucine transport(GO:0015820) |
| 0.2 | 0.5 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.2 | 0.7 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.2 | 0.5 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.2 | 0.8 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 1.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 0.8 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.6 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 3.1 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.4 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.4 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.4 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.1 | 0.4 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.1 | 0.8 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.5 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 2.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 1.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.5 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.4 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 1.0 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 1.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 2.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.6 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.4 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.5 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 | 3.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.3 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.9 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 0.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.8 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.4 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.6 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 1.0 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 1.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 1.5 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 1.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 1.5 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 1.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.6 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 6.0 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.1 | 0.5 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 1.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.4 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.9 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.3 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.1 | 1.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.3 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.1 | 1.5 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.4 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 0.3 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.7 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.5 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 2.6 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 0.4 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.2 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.2 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.1 | 0.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.4 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 0.5 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.8 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 2.5 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 0.4 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.4 | GO:0006231 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.1 | 0.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.5 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.4 | GO:0015853 | adenine transport(GO:0015853) |
| 0.1 | 1.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 2.4 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 | 2.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.1 | 0.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.4 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.5 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 1.6 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 2.5 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.2 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.1 | 2.3 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 1.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.3 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.1 | 0.3 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 1.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.7 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.2 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.6 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.4 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 2.1 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 1.8 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.5 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.4 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 1.8 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 1.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 3.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.7 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.6 | GO:0044351 | macropinocytosis(GO:0044351) phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.4 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 1.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.3 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 | 0.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.6 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 2.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.4 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.4 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.8 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 1.2 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.7 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.2 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 1.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.3 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.1 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.2 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 1.2 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.5 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.8 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.3 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 1.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.3 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.8 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.9 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 1.1 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.8 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 1.2 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.4 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.2 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 1.0 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.3 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.2 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.7 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.6 | GO:0035587 | purinergic receptor signaling pathway(GO:0035587) purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 2.1 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.4 | GO:2000647 | negative regulation of stem cell proliferation(GO:2000647) |
| 0.0 | 0.2 | GO:0044819 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) mitotic G1 DNA damage checkpoint(GO:0031571) G1 DNA damage checkpoint(GO:0044783) mitotic G1/S transition checkpoint(GO:0044819) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.5 | GO:0090314 | positive regulation of protein targeting to membrane(GO:0090314) |
| 0.0 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 1.0 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.3 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.5 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.0 | 0.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.5 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.0 | 0.4 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 1.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.7 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.0 | 0.4 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.7 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.3 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.4 | 1.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.3 | 2.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 1.6 | GO:0044393 | microspike(GO:0044393) |
| 0.3 | 1.8 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.3 | 0.8 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 1.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 0.6 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.2 | 0.6 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.2 | 1.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 0.7 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.1 | 0.4 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.4 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 1.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.4 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.1 | 1.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 3.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.7 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.1 | 2.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0071745 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.0 | 1.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 1.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 5.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 1.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.2 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 4.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.8 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 3.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 10.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 3.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 1.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 7.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 2.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 2.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.7 | 2.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.6 | 1.9 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.6 | 2.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.5 | 2.1 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.5 | 2.0 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.5 | 1.4 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.5 | 1.8 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.4 | 1.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.3 | 3.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.3 | 1.0 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.3 | 1.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.3 | 1.7 | GO:0070905 | serine binding(GO:0070905) |
| 0.3 | 0.8 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.3 | 1.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 0.7 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.2 | 2.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.7 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.2 | 0.9 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 2.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.9 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.2 | 1.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.9 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.2 | 0.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 0.6 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 0.6 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.2 | 1.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 2.7 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 1.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 1.0 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.2 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.5 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 0.7 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.2 | 0.5 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.2 | 1.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.6 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.6 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 1.0 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 2.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 1.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.6 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.1 | 0.4 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 1.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.9 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.4 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.1 | 1.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.5 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.4 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.4 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 0.3 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.1 | 1.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 2.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.5 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.6 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 2.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 1.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 2.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 2.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.2 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 0.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 3.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 3.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.2 | GO:0052830 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.1 | 0.2 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 0.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.6 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.2 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 4.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 1.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 2.3 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 2.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.0 | GO:0071617 | lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 1.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.5 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 2.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 2.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 3.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.0 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.4 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 2.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 7.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 4.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 3.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 2.1 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 2.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 5.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 3.4 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |