Project
Illumina Body Map 2
Navigation
Downloads
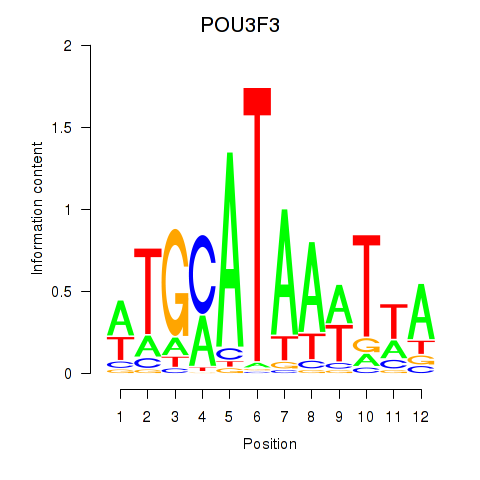
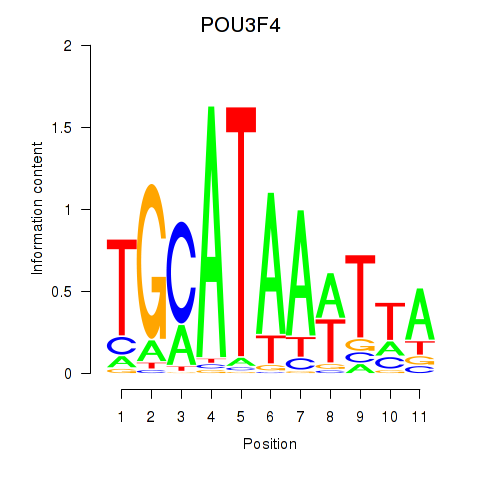
Results for POU3F3_POU3F4
Z-value: 1.60
Transcription factors associated with POU3F3_POU3F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU3F3
|
ENSG00000198914.2 | POU class 3 homeobox 3 |
|
POU3F4
|
ENSG00000196767.4 | POU class 3 homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU3F3 | hg19_v2_chr2_+_105471969_105471969 | 0.61 | 2.0e-04 | Click! |
| POU3F4 | hg19_v2_chrX_+_82763265_82763283 | 0.40 | 2.4e-02 | Click! |
Activity profile of POU3F3_POU3F4 motif
Sorted Z-values of POU3F3_POU3F4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_158142750 | 6.55 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr4_-_168155700 | 6.18 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_-_168155577 | 6.18 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr9_+_77230499 | 6.02 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr4_-_168155730 | 5.87 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr12_+_41221794 | 5.82 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr4_+_158141899 | 5.43 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr3_-_58613323 | 5.32 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr2_-_183387064 | 5.30 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr6_-_46459675 | 5.14 |
ENST00000306764.7
|
RCAN2
|
regulator of calcineurin 2 |
| chr8_-_91095099 | 4.89 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr3_+_35722487 | 4.77 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_35722844 | 4.75 |
ENST00000436702.1
ENST00000438071.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr15_+_76352178 | 4.63 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr5_-_160973649 | 4.43 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr3_+_158787041 | 4.35 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr11_-_26593649 | 4.07 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr2_-_166930131 | 3.99 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr20_+_58203664 | 3.93 |
ENST00000541461.1
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr10_-_128359074 | 3.91 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr17_+_3379284 | 3.88 |
ENST00000263080.2
|
ASPA
|
aspartoacylase |
| chr3_+_35722424 | 3.70 |
ENST00000396481.2
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr14_-_21270561 | 3.54 |
ENST00000412779.2
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr17_-_66951474 | 3.46 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr2_-_183387430 | 3.44 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_-_183387283 | 3.44 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr13_+_113698946 | 3.41 |
ENST00000397021.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr4_-_87281224 | 3.40 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr18_+_616711 | 3.40 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr11_-_26593677 | 3.37 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr16_-_21314360 | 3.36 |
ENST00000219599.3
ENST00000576703.1 |
CRYM
|
crystallin, mu |
| chr4_-_21950356 | 3.34 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr10_-_128359008 | 3.30 |
ENST00000488181.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr7_+_119913688 | 3.28 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr15_-_45670717 | 3.28 |
ENST00000558163.1
ENST00000558336.1 |
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr7_-_14880892 | 3.25 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr17_-_50236039 | 3.19 |
ENST00000451037.2
|
CA10
|
carbonic anhydrase X |
| chr5_-_11588907 | 3.15 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr5_-_11589131 | 3.12 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr3_-_100558953 | 3.11 |
ENST00000533795.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chrX_+_65382433 | 3.11 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr14_-_21270995 | 3.10 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr15_-_45670924 | 3.05 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr11_+_8040739 | 3.03 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr3_-_185826855 | 2.99 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr12_+_58003935 | 2.99 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr5_+_140753444 | 2.98 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr7_+_151653464 | 2.97 |
ENST00000431418.2
ENST00000392800.2 |
GALNTL5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr18_-_24722995 | 2.92 |
ENST00000581714.1
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr16_-_14109841 | 2.89 |
ENST00000576797.1
ENST00000575424.1 |
CTD-2135D7.5
|
CTD-2135D7.5 |
| chr2_-_183106641 | 2.83 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_+_140762268 | 2.81 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr10_+_24738355 | 2.81 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr4_+_88754113 | 2.79 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr12_+_41086297 | 2.78 |
ENST00000551295.2
|
CNTN1
|
contactin 1 |
| chr5_+_175288631 | 2.78 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr5_+_36608422 | 2.77 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr12_-_91573249 | 2.75 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr14_+_101297740 | 2.73 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr10_+_68685764 | 2.72 |
ENST00000361320.4
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3 |
| chr17_-_38821373 | 2.69 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr5_+_140529630 | 2.68 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr3_+_148508845 | 2.62 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr4_-_100575781 | 2.61 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
| chr14_-_61116168 | 2.56 |
ENST00000247182.6
|
SIX1
|
SIX homeobox 1 |
| chr16_-_10652993 | 2.47 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr12_-_10324716 | 2.43 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr1_-_151345159 | 2.42 |
ENST00000458566.1
ENST00000447402.3 ENST00000426705.2 ENST00000435071.1 ENST00000368868.5 |
SELENBP1
|
selenium binding protein 1 |
| chr11_-_56000737 | 2.42 |
ENST00000313264.4
|
OR5T2
|
olfactory receptor, family 5, subfamily T, member 2 |
| chr5_-_160279207 | 2.40 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr13_+_113699029 | 2.39 |
ENST00000423251.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr4_-_186570679 | 2.39 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_-_95657231 | 2.39 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chr5_+_125759140 | 2.34 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr3_-_196242233 | 2.34 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr1_+_209602609 | 2.33 |
ENST00000458250.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr9_+_137967268 | 2.33 |
ENST00000371799.4
ENST00000277415.11 |
OLFM1
|
olfactomedin 1 |
| chr4_+_158141843 | 2.29 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr2_+_40973618 | 2.24 |
ENST00000420187.1
|
AC007317.1
|
AC007317.1 |
| chr19_+_18726786 | 2.22 |
ENST00000594709.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr1_+_209602156 | 2.20 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr12_+_41221975 | 2.16 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chrX_-_72097698 | 2.15 |
ENST00000373530.1
|
DMRTC1
|
DMRT-like family C1 |
| chr21_+_17442799 | 2.12 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_-_44124019 | 2.12 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chrX_+_130192318 | 2.12 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr14_-_80697396 | 2.12 |
ENST00000557010.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chrX_+_72062617 | 2.08 |
ENST00000440247.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr15_+_69857515 | 2.07 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr5_+_125758813 | 2.04 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr9_+_34458771 | 2.04 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr5_+_125758865 | 2.03 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr11_-_40314652 | 2.01 |
ENST00000527150.1
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr8_-_67090825 | 1.97 |
ENST00000276571.3
|
CRH
|
corticotropin releasing hormone |
| chr1_+_170632250 | 1.96 |
ENST00000367760.3
|
PRRX1
|
paired related homeobox 1 |
| chr18_+_76740189 | 1.95 |
ENST00000537592.2
ENST00000575389.2 |
SALL3
|
spalt-like transcription factor 3 |
| chr8_-_38008783 | 1.94 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr2_+_166095898 | 1.93 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr21_+_17443521 | 1.92 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr15_-_76304731 | 1.92 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr10_-_128975273 | 1.92 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr11_-_102709441 | 1.91 |
ENST00000434103.1
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr12_+_41086215 | 1.89 |
ENST00000547702.1
ENST00000551424.1 |
CNTN1
|
contactin 1 |
| chr15_-_76352069 | 1.89 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chrX_-_32173579 | 1.87 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr3_-_149375783 | 1.87 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr8_-_36636676 | 1.85 |
ENST00000524132.1
ENST00000519451.1 |
RP11-962G15.1
|
RP11-962G15.1 |
| chr6_+_127898312 | 1.79 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr10_+_29577974 | 1.78 |
ENST00000375500.3
|
LYZL1
|
lysozyme-like 1 |
| chr3_-_74570291 | 1.77 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr5_+_140710061 | 1.77 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr1_-_169703203 | 1.76 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr3_+_159943362 | 1.76 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr4_-_52883786 | 1.75 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr4_+_9446156 | 1.73 |
ENST00000334879.1
|
DEFB131
|
defensin, beta 131 |
| chr17_-_10452929 | 1.73 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr2_+_210444142 | 1.71 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr19_-_44123734 | 1.69 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr15_+_71228826 | 1.68 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr10_-_48806939 | 1.67 |
ENST00000374233.3
ENST00000507417.1 ENST00000512321.1 ENST00000395660.2 ENST00000374235.2 ENST00000395661.3 |
PTPN20B
|
protein tyrosine phosphatase, non-receptor type 20B |
| chr7_-_229557 | 1.67 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr15_-_74658493 | 1.66 |
ENST00000419019.2
ENST00000569662.1 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr12_-_91574142 | 1.63 |
ENST00000547937.1
|
DCN
|
decorin |
| chr16_-_745946 | 1.62 |
ENST00000562563.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr5_+_140602904 | 1.61 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr9_+_34458851 | 1.61 |
ENST00000545019.1
|
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr3_+_125694347 | 1.61 |
ENST00000505382.1
ENST00000511082.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr7_-_15014398 | 1.60 |
ENST00000437998.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr11_-_88799113 | 1.60 |
ENST00000393294.3
|
GRM5
|
glutamate receptor, metabotropic 5 |
| chr15_-_74658519 | 1.57 |
ENST00000450547.1
ENST00000358632.4 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr7_-_100026280 | 1.56 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr4_+_30723003 | 1.53 |
ENST00000543491.1
|
PCDH7
|
protocadherin 7 |
| chr5_-_133702761 | 1.52 |
ENST00000521118.1
ENST00000265334.4 ENST00000435211.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr2_+_239335449 | 1.52 |
ENST00000264607.4
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr1_+_53925063 | 1.51 |
ENST00000371445.3
|
DMRTB1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr1_+_28099683 | 1.50 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr2_-_239140276 | 1.48 |
ENST00000334973.4
|
AC016757.3
|
Protein LOC151174 |
| chr5_+_140782351 | 1.47 |
ENST00000573521.1
|
PCDHGA9
|
protocadherin gamma subfamily A, 9 |
| chr7_-_16505440 | 1.46 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr5_+_140474181 | 1.46 |
ENST00000194155.4
|
PCDHB2
|
protocadherin beta 2 |
| chr11_-_57004658 | 1.44 |
ENST00000606794.1
|
APLNR
|
apelin receptor |
| chr15_-_49255632 | 1.42 |
ENST00000332408.4
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr13_+_109248500 | 1.42 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr18_-_25739260 | 1.41 |
ENST00000413878.1
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr12_-_91573132 | 1.41 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr16_+_76311169 | 1.40 |
ENST00000307431.8
ENST00000377504.4 |
CNTNAP4
|
contactin associated protein-like 4 |
| chr2_-_101034070 | 1.39 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr3_-_73610759 | 1.39 |
ENST00000466780.1
|
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr1_+_28099700 | 1.38 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chrX_-_112084043 | 1.36 |
ENST00000304758.1
|
AMOT
|
angiomotin |
| chr18_+_616672 | 1.35 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr5_+_140174429 | 1.35 |
ENST00000520672.2
ENST00000378132.1 ENST00000526136.1 |
PCDHA2
|
protocadherin alpha 2 |
| chr3_-_78719376 | 1.35 |
ENST00000495961.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr9_+_137967366 | 1.34 |
ENST00000252854.4
|
OLFM1
|
olfactomedin 1 |
| chr17_-_66951382 | 1.34 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr15_-_30113676 | 1.34 |
ENST00000400011.2
|
TJP1
|
tight junction protein 1 |
| chr12_-_91573316 | 1.33 |
ENST00000393155.1
|
DCN
|
decorin |
| chr12_-_120241187 | 1.33 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr6_+_168418553 | 1.33 |
ENST00000354419.2
ENST00000351261.3 |
KIF25
|
kinesin family member 25 |
| chrX_-_18690210 | 1.33 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr10_-_75423560 | 1.31 |
ENST00000606523.1
|
SYNPO2L
|
synaptopodin 2-like |
| chr3_-_62359180 | 1.30 |
ENST00000283268.3
|
FEZF2
|
FEZ family zinc finger 2 |
| chr2_-_239140011 | 1.29 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr1_-_216978709 | 1.29 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr1_-_193155729 | 1.28 |
ENST00000367434.4
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr21_+_17443434 | 1.28 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr6_-_52859046 | 1.28 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr15_-_90892669 | 1.27 |
ENST00000412799.2
|
GABARAPL3
|
GABA(A) receptors associated protein like 3, pseudogene |
| chr22_+_30476163 | 1.27 |
ENST00000336726.6
|
HORMAD2
|
HORMA domain containing 2 |
| chr3_+_173116225 | 1.27 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr16_-_30064244 | 1.27 |
ENST00000571269.1
ENST00000561666.1 |
FAM57B
|
family with sequence similarity 57, member B |
| chrX_-_80457385 | 1.27 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr12_-_71148413 | 1.27 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr5_+_126984710 | 1.26 |
ENST00000379445.3
|
CTXN3
|
cortexin 3 |
| chr18_-_48346415 | 1.25 |
ENST00000431965.2
ENST00000436348.2 |
MRO
|
maestro |
| chr14_-_47351391 | 1.25 |
ENST00000399222.3
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr3_-_73483055 | 1.25 |
ENST00000479530.1
|
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr3_+_53528659 | 1.24 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr2_+_189839046 | 1.24 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chrX_-_154255143 | 1.23 |
ENST00000453950.1
ENST00000423959.1 |
F8
|
coagulation factor VIII, procoagulant component |
| chr8_-_124749609 | 1.23 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr12_-_71148357 | 1.23 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr20_-_21378666 | 1.23 |
ENST00000351817.4
|
NKX2-4
|
NK2 homeobox 4 |
| chr8_-_119634141 | 1.22 |
ENST00000409003.4
ENST00000526328.1 ENST00000314727.4 ENST00000526765.1 |
SAMD12
|
sterile alpha motif domain containing 12 |
| chr2_+_210443993 | 1.21 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr9_-_15472730 | 1.21 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr4_-_20985632 | 1.20 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr10_-_124459284 | 1.20 |
ENST00000432000.1
ENST00000329446.4 |
C10orf120
|
chromosome 10 open reading frame 120 |
| chr3_+_189507523 | 1.19 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chrX_+_28605516 | 1.19 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr5_-_78809950 | 1.19 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr5_-_42811986 | 1.19 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr5_+_149980622 | 1.19 |
ENST00000394243.1
|
SYNPO
|
synaptopodin |
| chr4_-_176708533 | 1.17 |
ENST00000507520.1
|
GPM6A
|
glycoprotein M6A |
| chr8_-_65711310 | 1.17 |
ENST00000310193.3
|
CYP7B1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chrY_-_20935572 | 1.17 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr4_-_152149033 | 1.16 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr3_-_33686925 | 1.15 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr4_+_114066764 | 1.15 |
ENST00000511380.1
|
ANK2
|
ankyrin 2, neuronal |
| chr1_+_113009163 | 1.15 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU3F3_POU3F4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 1.3 | 3.9 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 1.1 | 25.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.8 | 3.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.8 | 15.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.7 | 4.9 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.7 | 2.8 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.7 | 2.0 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) positive regulation of corticosterone secretion(GO:2000854) |
| 0.6 | 2.6 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.6 | 6.0 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.6 | 2.9 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.5 | 1.9 | GO:0018963 | insecticide metabolic process(GO:0017143) phthalate metabolic process(GO:0018963) |
| 0.5 | 1.4 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.4 | 1.3 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.4 | 4.0 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.4 | 2.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.3 | 3.8 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 2.8 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 1.3 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.3 | 1.3 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.3 | 5.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 1.3 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.3 | 4.8 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.3 | 1.5 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 3.4 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.3 | 3.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.3 | 0.8 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.3 | 3.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.3 | 5.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.3 | 1.3 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.3 | 2.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 3.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 1.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 3.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 14.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 0.6 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 0.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 2.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 1.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 1.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.2 | 4.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 14.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.2 | 1.4 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 3.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 1.4 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.2 | 0.6 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 1.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 3.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.5 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.1 | 5.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.5 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 1.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 2.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 4.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 2.7 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 1.1 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 0.8 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 1.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.6 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 2.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 1.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 2.0 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 1.9 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.3 | GO:1904956 | neural crest cell fate commitment(GO:0014034) cellular response to heparin(GO:0071504) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 2.8 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 1.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 2.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 1.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 1.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 1.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.4 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 4.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 3.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 1.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.7 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.1 | 3.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 1.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 1.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 1.5 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 6.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 2.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.9 | GO:1903209 | positive regulation of oxidative stress-induced cell death(GO:1903209) |
| 0.1 | 0.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 2.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 6.7 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 7.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 12.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 4.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.5 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.1 | 0.8 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.1 | 1.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.3 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 1.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 1.4 | GO:0048863 | stem cell differentiation(GO:0048863) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 12.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 2.9 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 3.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 1.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.8 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 3.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.8 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.9 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.2 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 1.3 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 3.5 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.5 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 1.3 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.5 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.6 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 1.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 6.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.3 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.2 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.0 | 1.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.8 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.9 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 3.0 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 1.1 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 2.8 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 4.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 1.7 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 1.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.5 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) |
| 0.0 | 0.7 | GO:0033260 | DNA replication initiation(GO:0006270) nuclear DNA replication(GO:0033260) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 2.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 14.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.5 | 7.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.4 | 1.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.3 | 3.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.3 | 2.8 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.3 | 2.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 6.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 2.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 1.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.9 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 3.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 4.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 4.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 3.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.7 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 3.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.6 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.7 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 5.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 7.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 5.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 11.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 21.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 3.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 3.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 6.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 3.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 18.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.7 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.1 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 2.0 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 3.4 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 8.6 | GO:0005813 | centrosome(GO:0005813) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 15.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 2.1 | 6.3 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 1.1 | 3.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.9 | 6.6 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.8 | 3.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.8 | 14.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.7 | 5.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 4.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.6 | 18.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 4.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.5 | 2.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.5 | 2.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 1.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.4 | 2.9 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.4 | 1.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.4 | 3.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 3.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 4.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 4.8 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.4 | 3.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 3.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 1.6 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.3 | 2.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.2 | 0.6 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 6.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 3.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 6.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 1.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 4.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 2.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.5 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.2 | 0.8 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 1.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 0.6 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.2 | 3.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 13.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 0.6 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 3.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 2.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 5.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.8 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.1 | 0.6 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 2.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 1.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 0.4 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 2.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 7.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.6 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 2.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.9 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 1.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 1.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 2.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 2.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 1.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 2.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 16.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 6.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 9.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 3.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 3.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 4.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.5 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.9 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 25.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 15.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 8.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 11.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 4.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 4.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 3.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 3.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.3 | 15.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 10.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 10.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 2.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 7.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 3.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 4.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 4.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 5.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 4.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 5.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 4.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 7.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 3.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 3.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 2.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 3.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 5.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 3.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 2.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |