Project
Illumina Body Map 2
Navigation
Downloads
Results for POU4F1_POU4F3
Z-value: 1.02
Transcription factors associated with POU4F1_POU4F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
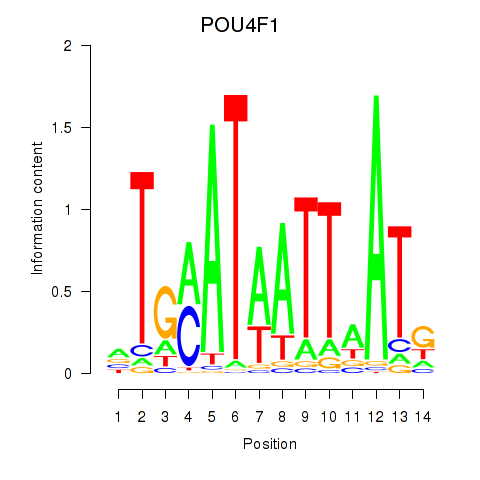
POU4F1
|
ENSG00000152192.6 | POU class 4 homeobox 1 |
|
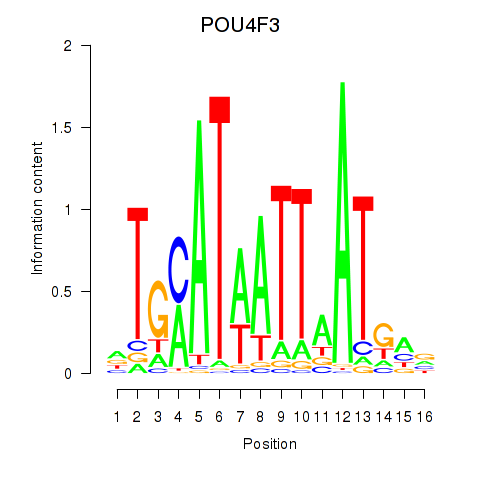
POU4F3
|
ENSG00000091010.4 | POU class 4 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU4F1 | hg19_v2_chr13_-_79177673_79177701 | 0.28 | 1.2e-01 | Click! |
| POU4F3 | hg19_v2_chr5_+_145718587_145718607 | 0.10 | 5.8e-01 | Click! |
Activity profile of POU4F1_POU4F3 motif
Sorted Z-values of POU4F1_POU4F3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_38821373 | 3.92 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr1_+_50574585 | 3.79 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr4_+_74269956 | 3.68 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr18_-_31803435 | 3.24 |
ENST00000589544.1
ENST00000269185.4 ENST00000261592.5 |
NOL4
|
nucleolar protein 4 |
| chr4_-_72649763 | 3.08 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr2_-_88285309 | 3.07 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr12_+_41221794 | 3.02 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr12_+_51985001 | 2.94 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr3_+_68055366 | 2.78 |
ENST00000496687.1
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| chr22_+_30476163 | 2.63 |
ENST00000336726.6
|
HORMAD2
|
HORMA domain containing 2 |
| chr14_-_65409438 | 2.58 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr6_-_136847099 | 2.53 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr12_+_51984657 | 2.47 |
ENST00000550891.1
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chrX_-_138724677 | 2.43 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chrX_-_138724994 | 2.37 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr9_+_34458771 | 2.28 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr3_+_35722487 | 2.26 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr6_-_136847610 | 2.26 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr19_+_17865011 | 2.23 |
ENST00000596462.1
ENST00000596865.1 ENST00000598960.1 ENST00000539407.1 |
FCHO1
|
FCH domain only 1 |
| chr3_-_52860850 | 2.22 |
ENST00000441637.2
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr12_+_79357815 | 2.12 |
ENST00000547046.1
|
SYT1
|
synaptotagmin I |
| chr2_+_87135076 | 2.08 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr14_-_65409502 | 1.89 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr9_+_34458851 | 1.86 |
ENST00000545019.1
|
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr3_+_35722844 | 1.84 |
ENST00000436702.1
ENST00000438071.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_35722424 | 1.83 |
ENST00000396481.2
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chrX_-_106146547 | 1.79 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr18_-_31803169 | 1.78 |
ENST00000590712.1
|
NOL4
|
nucleolar protein 4 |
| chr18_+_76740189 | 1.78 |
ENST00000537592.2
ENST00000575389.2 |
SALL3
|
spalt-like transcription factor 3 |
| chr22_+_30477000 | 1.75 |
ENST00000403975.1
|
HORMAD2
|
HORMA domain containing 2 |
| chr12_+_41221975 | 1.75 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr3_+_46742823 | 1.71 |
ENST00000326431.3
|
TMIE
|
transmembrane inner ear |
| chr19_-_36304201 | 1.71 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr1_+_63063152 | 1.70 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chr2_+_185463093 | 1.61 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr16_-_51185172 | 1.55 |
ENST00000251020.4
|
SALL1
|
spalt-like transcription factor 1 |
| chr11_-_35441524 | 1.55 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr16_-_51185149 | 1.51 |
ENST00000566102.1
ENST00000541611.1 |
SALL1
|
spalt-like transcription factor 1 |
| chr11_-_35441597 | 1.50 |
ENST00000395753.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr12_+_49740700 | 1.49 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr14_-_101036119 | 1.47 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr5_+_139739772 | 1.47 |
ENST00000506757.2
ENST00000230993.6 ENST00000506545.1 ENST00000432095.2 ENST00000507527.1 |
SLC4A9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr3_-_194072019 | 1.41 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr17_-_50236039 | 1.34 |
ENST00000451037.2
|
CA10
|
carbonic anhydrase X |
| chr17_+_18012020 | 1.32 |
ENST00000205890.5
|
MYO15A
|
myosin XVA |
| chr16_+_24621546 | 1.31 |
ENST00000566108.1
|
CTD-2540M10.1
|
CTD-2540M10.1 |
| chr21_-_37914898 | 1.27 |
ENST00000399136.1
|
CLDN14
|
claudin 14 |
| chr2_-_209054709 | 1.26 |
ENST00000449053.1
ENST00000451346.1 ENST00000341287.4 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr10_-_128359008 | 1.25 |
ENST00000488181.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr12_-_9885888 | 1.24 |
ENST00000327839.3
|
CLECL1
|
C-type lectin-like 1 |
| chr19_+_19639704 | 1.21 |
ENST00000514277.4
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr19_+_19639670 | 1.21 |
ENST00000436027.5
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr11_-_7698453 | 1.16 |
ENST00000524608.1
|
CYB5R2
|
cytochrome b5 reductase 2 |
| chr17_-_10741762 | 1.16 |
ENST00000580256.2
|
PIRT
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr2_+_182850551 | 1.14 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_+_179318295 | 1.11 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr19_-_13734804 | 1.11 |
ENST00000574974.1
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr15_+_58724184 | 1.09 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr4_-_21546272 | 1.07 |
ENST00000509207.1
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr9_-_123812542 | 1.07 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr1_+_149239529 | 1.07 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr1_+_101003687 | 1.04 |
ENST00000315033.4
|
GPR88
|
G protein-coupled receptor 88 |
| chr1_-_108231101 | 1.04 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr1_+_160336851 | 1.04 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr9_-_74675521 | 1.00 |
ENST00000377024.3
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr4_+_159236462 | 0.99 |
ENST00000460056.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr2_-_183387283 | 0.97 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr12_+_100750846 | 0.95 |
ENST00000323346.5
|
SLC17A8
|
solute carrier family 17 (vesicular glutamate transporter), member 8 |
| chr10_-_128359074 | 0.95 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr5_-_13944652 | 0.93 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr11_+_58706944 | 0.90 |
ENST00000532726.1
|
GLYATL1
|
glycine-N-acyltransferase-like 1 |
| chrX_-_142605301 | 0.90 |
ENST00000370503.2
|
SPANXN3
|
SPANX family, member N3 |
| chr15_+_76352178 | 0.89 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr22_-_30970560 | 0.88 |
ENST00000402369.1
ENST00000406361.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr8_-_110986918 | 0.88 |
ENST00000297404.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr11_-_83393429 | 0.88 |
ENST00000426717.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr2_+_179317994 | 0.87 |
ENST00000375129.4
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr6_-_10115007 | 0.87 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr11_+_98891797 | 0.87 |
ENST00000527185.1
ENST00000528682.1 ENST00000524871.1 |
CNTN5
|
contactin 5 |
| chr17_-_72855989 | 0.84 |
ENST00000293190.5
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
| chr20_+_30555805 | 0.82 |
ENST00000562532.2
|
XKR7
|
XK, Kell blood group complex subunit-related family, member 7 |
| chr17_-_56492989 | 0.80 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr5_-_135290705 | 0.79 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr19_-_51141196 | 0.78 |
ENST00000338916.4
|
SYT3
|
synaptotagmin III |
| chr1_-_197036364 | 0.78 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr11_-_83393457 | 0.77 |
ENST00000404783.3
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chrX_-_72097698 | 0.77 |
ENST00000373530.1
|
DMRTC1
|
DMRT-like family C1 |
| chr2_-_183387430 | 0.77 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_+_47603109 | 0.77 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr3_-_185826855 | 0.75 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chrX_+_72062802 | 0.74 |
ENST00000373533.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr10_-_128210005 | 0.74 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr2_+_133174147 | 0.74 |
ENST00000329321.3
|
GPR39
|
G protein-coupled receptor 39 |
| chrX_+_72062617 | 0.74 |
ENST00000440247.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr5_-_11589131 | 0.72 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr7_-_107880508 | 0.72 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr18_-_70305745 | 0.72 |
ENST00000581073.1
|
CBLN2
|
cerebellin 2 precursor |
| chr11_-_83393303 | 0.70 |
ENST00000398304.1
ENST00000420775.2 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr17_+_28268623 | 0.69 |
ENST00000394835.3
ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr18_+_47088401 | 0.69 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chr2_-_183387064 | 0.67 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_-_100026280 | 0.67 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr19_+_54466179 | 0.65 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr11_-_26593649 | 0.65 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr5_-_11588907 | 0.63 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr12_-_120241187 | 0.63 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr18_-_24443151 | 0.62 |
ENST00000440832.3
|
AQP4
|
aquaporin 4 |
| chr1_-_248845629 | 0.60 |
ENST00000342623.3
|
OR14I1
|
olfactory receptor, family 14, subfamily I, member 1 |
| chr22_-_38506619 | 0.60 |
ENST00000332536.5
ENST00000381669.3 |
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr2_+_87144738 | 0.60 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr12_+_81664424 | 0.59 |
ENST00000549161.1
ENST00000550138.1 |
RP11-121G22.3
|
RP11-121G22.3 |
| chr4_-_69817481 | 0.59 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr19_-_39402798 | 0.58 |
ENST00000571838.1
|
CTC-360G5.1
|
coiled-coil glutamate-rich protein 2 |
| chr9_-_16253112 | 0.58 |
ENST00000380683.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr2_+_197577841 | 0.56 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr11_-_88799113 | 0.55 |
ENST00000393294.3
|
GRM5
|
glutamate receptor, metabotropic 5 |
| chr7_+_99195677 | 0.55 |
ENST00000431679.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr22_-_30970498 | 0.53 |
ENST00000431313.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr7_-_15014398 | 0.50 |
ENST00000437998.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr6_-_127840021 | 0.50 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr6_-_76782371 | 0.50 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr11_-_69587718 | 0.49 |
ENST00000602104.1
|
AP001888.1
|
Uncharacterized protein |
| chr19_-_55677920 | 0.47 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr19_+_56368803 | 0.45 |
ENST00000587891.1
|
NLRP4
|
NLR family, pyrin domain containing 4 |
| chr9_+_35673853 | 0.45 |
ENST00000378357.4
|
CA9
|
carbonic anhydrase IX |
| chr3_-_33686925 | 0.44 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_-_26108355 | 0.44 |
ENST00000338379.4
|
HIST1H1T
|
histone cluster 1, H1t |
| chr12_+_110011571 | 0.43 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr14_+_101297740 | 0.41 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr3_-_74570291 | 0.41 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr1_-_222014008 | 0.41 |
ENST00000431729.1
|
RP11-191N8.2
|
RP11-191N8.2 |
| chr2_+_182850743 | 0.41 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr5_-_135290651 | 0.40 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr13_-_20110902 | 0.38 |
ENST00000390680.2
ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr4_-_103998439 | 0.38 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr2_+_65663812 | 0.38 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr17_+_37783197 | 0.38 |
ENST00000582680.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr7_-_35013217 | 0.38 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr10_+_90346519 | 0.38 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chr11_+_86085778 | 0.37 |
ENST00000354755.1
ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81
|
coiled-coil domain containing 81 |
| chr2_+_191221240 | 0.37 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr13_-_70682590 | 0.37 |
ENST00000377844.4
|
KLHL1
|
kelch-like family member 1 |
| chr22_+_17956618 | 0.36 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr1_-_47017199 | 0.36 |
ENST00000481882.2
|
KNCN
|
kinocilin |
| chr6_-_127840336 | 0.36 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr22_+_43011247 | 0.35 |
ENST00000602478.1
|
RNU12
|
RNA, U12 small nuclear |
| chr9_-_21166659 | 0.34 |
ENST00000380225.1
|
IFNA21
|
interferon, alpha 21 |
| chr2_-_26781521 | 0.34 |
ENST00000403946.3
ENST00000272371.2 |
OTOF
|
otoferlin |
| chr11_+_22696314 | 0.34 |
ENST00000532398.1
ENST00000433790.1 |
GAS2
|
growth arrest-specific 2 |
| chr3_-_33686743 | 0.34 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr1_-_154580616 | 0.34 |
ENST00000368474.4
|
ADAR
|
adenosine deaminase, RNA-specific |
| chr10_-_128975273 | 0.34 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr17_+_37782955 | 0.34 |
ENST00000580825.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr17_+_37783453 | 0.33 |
ENST00000579000.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr20_-_29978383 | 0.33 |
ENST00000339144.3
ENST00000376321.3 |
DEFB119
|
defensin, beta 119 |
| chr22_-_39268308 | 0.33 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr1_+_161691353 | 0.32 |
ENST00000367948.2
|
FCRLB
|
Fc receptor-like B |
| chr4_+_68424434 | 0.32 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr14_+_22580233 | 0.31 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr2_+_47454054 | 0.31 |
ENST00000426892.1
|
AC106869.2
|
AC106869.2 |
| chr17_-_37123646 | 0.30 |
ENST00000378079.2
|
FBXO47
|
F-box protein 47 |
| chr10_+_104614008 | 0.30 |
ENST00000369883.3
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr6_+_126102292 | 0.30 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr12_-_23737534 | 0.30 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr11_-_26593677 | 0.30 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr4_+_146601356 | 0.29 |
ENST00000438731.1
ENST00000511965.1 |
C4orf51
|
chromosome 4 open reading frame 51 |
| chr9_-_107361788 | 0.29 |
ENST00000374779.2
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5 |
| chr17_+_37783170 | 0.29 |
ENST00000254079.4
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr2_+_40973618 | 0.28 |
ENST00000420187.1
|
AC007317.1
|
AC007317.1 |
| chr4_-_122085469 | 0.27 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr4_-_16085314 | 0.27 |
ENST00000510224.1
|
PROM1
|
prominin 1 |
| chr3_-_167371704 | 0.27 |
ENST00000488012.1
|
WDR49
|
WD repeat domain 49 |
| chr12_+_130554803 | 0.27 |
ENST00000535487.1
|
RP11-474D1.2
|
RP11-474D1.2 |
| chr3_-_167371740 | 0.26 |
ENST00000466760.1
ENST00000479765.1 |
WDR49
|
WD repeat domain 49 |
| chr10_-_112678904 | 0.26 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr14_+_22978168 | 0.26 |
ENST00000390505.1
|
TRAJ32
|
T cell receptor alpha joining 32 |
| chr16_-_67217844 | 0.26 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr1_-_95538492 | 0.26 |
ENST00000370205.5
|
ALG14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr11_-_111649074 | 0.26 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr19_-_23941639 | 0.25 |
ENST00000395385.3
ENST00000531570.1 ENST00000528059.1 |
ZNF681
|
zinc finger protein 681 |
| chr10_+_135207598 | 0.25 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr2_-_183106641 | 0.24 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr6_-_2634820 | 0.24 |
ENST00000296847.3
|
C6orf195
|
chromosome 6 open reading frame 195 |
| chr4_-_16085340 | 0.24 |
ENST00000508167.1
|
PROM1
|
prominin 1 |
| chr19_-_46580352 | 0.24 |
ENST00000601672.1
|
IGFL4
|
IGF-like family member 4 |
| chr16_-_52061283 | 0.24 |
ENST00000566314.1
|
C16orf97
|
chromosome 16 open reading frame 97 |
| chr3_-_185826718 | 0.23 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr10_-_112678692 | 0.23 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr5_-_39270725 | 0.23 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr9_-_4666421 | 0.23 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr13_+_113698946 | 0.23 |
ENST00000397021.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr10_+_135207623 | 0.23 |
ENST00000317502.6
ENST00000432508.3 |
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr2_+_169757750 | 0.23 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr9_-_4666337 | 0.22 |
ENST00000381890.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr20_+_57875758 | 0.22 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr10_+_104613980 | 0.21 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr13_+_46039037 | 0.21 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr19_-_55677999 | 0.21 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr6_-_52859046 | 0.20 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr6_-_161695074 | 0.20 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU4F1_POU4F3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.5 | 3.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.4 | 4.4 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.4 | 2.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.3 | 1.8 | GO:0001757 | somite specification(GO:0001757) |
| 0.3 | 0.8 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.3 | 3.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 1.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 5.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 0.7 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.2 | 1.7 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 5.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 0.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.6 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 2.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.0 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 1.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.3 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.8 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.1 | 0.5 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 1.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 3.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 5.6 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 1.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.8 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.2 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 2.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.3 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.0 | 0.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 1.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 1.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.7 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 3.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 2.1 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 1.9 | GO:0046710 | GDP metabolic process(GO:0046710) receptor localization to synapse(GO:0097120) |
| 0.0 | 4.8 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.4 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 5.1 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.2 | GO:0014826 | cellular magnesium ion homeostasis(GO:0010961) vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 1.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 5.7 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.6 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.5 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 1.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 3.9 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 1.0 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 5.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.2 | GO:0042330 | chemotaxis(GO:0006935) taxis(GO:0042330) |
| 0.0 | 0.6 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 3.2 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0050777 | negative regulation of immune response(GO:0050777) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 1.3 | GO:0007420 | brain development(GO:0007420) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) nuclear pore complex assembly(GO:0051292) endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.2 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.3 | 1.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 2.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 5.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 3.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 4.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 2.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 3.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 1.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 6.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 4.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 4.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 2.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 9.2 | GO:0043025 | neuronal cell body(GO:0043025) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.8 | 2.4 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.4 | 1.7 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.4 | 2.7 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 3.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 1.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.2 | 0.9 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.2 | 2.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 4.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 5.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 3.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 4.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.4 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.4 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 1.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.4 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 3.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.8 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 1.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 0.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.6 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 1.0 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 2.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 1.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 5.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 1.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 5.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 4.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 3.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 2.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 6.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 4.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 3.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 5.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 4.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.8 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |