Project
Illumina Body Map 2
Navigation
Downloads
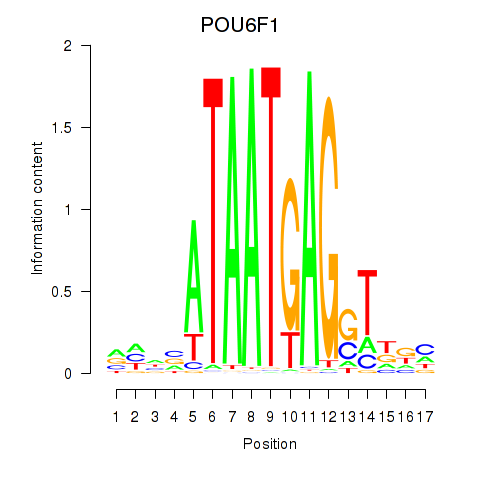
Results for POU6F1
Z-value: 0.76
Transcription factors associated with POU6F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F1
|
ENSG00000184271.11 | POU class 6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU6F1 | hg19_v2_chr12_-_51611477_51611612 | -0.14 | 4.3e-01 | Click! |
Activity profile of POU6F1 motif
Sorted Z-values of POU6F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_49297899 | 2.61 |
ENST00000552942.1
ENST00000320516.4 |
CCDC65
|
coiled-coil domain containing 65 |
| chr7_+_142012967 | 2.51 |
ENST00000390357.3
|
TRBV4-1
|
T cell receptor beta variable 4-1 |
| chr3_+_46395219 | 2.50 |
ENST00000445132.2
ENST00000292301.4 |
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr1_-_47655686 | 2.18 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr7_+_142045246 | 1.90 |
ENST00000390392.3
|
TRBV4-2
|
T cell receptor beta variable 4-2 |
| chr6_+_50786414 | 1.72 |
ENST00000344788.3
ENST00000393655.3 ENST00000263046.4 |
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr19_+_10959043 | 1.56 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr19_-_35719609 | 1.55 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187, member B |
| chr12_+_49297887 | 1.43 |
ENST00000266984.5
|
CCDC65
|
coiled-coil domain containing 65 |
| chr3_+_46395579 | 1.41 |
ENST00000421659.1
|
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr18_+_52495426 | 1.35 |
ENST00000262094.5
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr14_+_22689792 | 1.34 |
ENST00000390462.1
|
TRAV35
|
T cell receptor alpha variable 35 |
| chr6_+_42123141 | 1.31 |
ENST00000418175.1
ENST00000541991.1 ENST00000053469.4 ENST00000394237.1 ENST00000372963.1 |
GUCA1A
RP1-139D8.6
|
guanylate cyclase activator 1A (retina) RP1-139D8.6 |
| chr9_-_113100088 | 1.26 |
ENST00000374510.4
ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa) |
| chr7_+_80267949 | 1.21 |
ENST00000482059.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr4_+_40201954 | 1.20 |
ENST00000511121.1
|
RHOH
|
ras homolog family member H |
| chr11_+_12302492 | 1.19 |
ENST00000533534.1
|
MICALCL
|
MICAL C-terminal like |
| chr1_-_222014008 | 1.19 |
ENST00000431729.1
|
RP11-191N8.2
|
RP11-191N8.2 |
| chr18_+_61554932 | 1.13 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr20_+_59654146 | 1.02 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr1_-_106161540 | 1.02 |
ENST00000420901.1
ENST00000610126.1 ENST00000435253.2 |
RP11-251P6.1
|
RP11-251P6.1 |
| chr14_+_39944025 | 1.01 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr19_-_3786253 | 1.01 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr7_+_80267973 | 0.98 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr19_-_3786354 | 0.97 |
ENST00000395040.2
ENST00000310132.6 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr11_-_2323089 | 0.96 |
ENST00000456145.2
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr22_-_32767017 | 0.96 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr1_-_197115818 | 0.95 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr16_+_15744078 | 0.93 |
ENST00000396354.1
ENST00000570727.1 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr11_+_2323349 | 0.92 |
ENST00000381121.3
|
TSPAN32
|
tetraspanin 32 |
| chr15_+_45028719 | 0.92 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chr11_+_2323236 | 0.90 |
ENST00000182290.4
|
TSPAN32
|
tetraspanin 32 |
| chr11_-_107729504 | 0.89 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr15_+_45028753 | 0.87 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chr11_-_107729287 | 0.87 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr16_-_87350970 | 0.87 |
ENST00000567970.1
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chr12_-_57443886 | 0.84 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr20_-_58515344 | 0.84 |
ENST00000370996.3
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D |
| chr13_+_30510003 | 0.82 |
ENST00000400540.1
|
LINC00544
|
long intergenic non-protein coding RNA 544 |
| chr15_+_80351910 | 0.82 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr3_-_48659193 | 0.82 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr11_+_71544246 | 0.78 |
ENST00000328698.1
|
DEFB108B
|
defensin, beta 108B |
| chr6_-_74231444 | 0.78 |
ENST00000331523.2
ENST00000356303.2 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr2_+_191045656 | 0.76 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr20_+_58515417 | 0.76 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr12_+_54892550 | 0.71 |
ENST00000545638.2
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr1_-_243326612 | 0.66 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr1_-_153123345 | 0.66 |
ENST00000368748.4
|
SPRR2G
|
small proline-rich protein 2G |
| chr2_-_99917639 | 0.64 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr12_+_15699286 | 0.64 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr6_-_111136299 | 0.63 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr17_+_73539339 | 0.62 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr15_+_45028520 | 0.61 |
ENST00000329464.4
|
TRIM69
|
tripartite motif containing 69 |
| chr11_-_107729887 | 0.59 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_+_130692778 | 0.58 |
ENST00000513875.1
ENST00000508724.1 |
RP11-422J15.1
|
RP11-422J15.1 |
| chr8_+_118147498 | 0.58 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr22_-_22307199 | 0.57 |
ENST00000397495.4
ENST00000263212.5 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr6_-_110501126 | 0.56 |
ENST00000368938.1
|
WASF1
|
WAS protein family, member 1 |
| chr15_+_80351977 | 0.55 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr7_-_30008849 | 0.54 |
ENST00000409497.1
|
SCRN1
|
secernin 1 |
| chr16_-_87351022 | 0.52 |
ENST00000253461.4
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chr12_+_102513950 | 0.51 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chr19_+_52076425 | 0.51 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr18_-_73967160 | 0.49 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr15_+_74421961 | 0.49 |
ENST00000565159.1
ENST00000567206.1 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr13_-_52027134 | 0.48 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr6_-_110501200 | 0.46 |
ENST00000392586.1
ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1
|
WAS protein family, member 1 |
| chr8_+_78311303 | 0.46 |
ENST00000518706.2
|
RP11-38H17.1
|
RP11-38H17.1 |
| chr1_+_50571949 | 0.46 |
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chrX_-_122756660 | 0.45 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr5_+_55147205 | 0.44 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr19_+_45116921 | 0.44 |
ENST00000402988.1
|
IGSF23
|
immunoglobulin superfamily, member 23 |
| chr6_+_29274403 | 0.44 |
ENST00000377160.2
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1 |
| chr6_+_23337918 | 0.43 |
ENST00000431001.1
|
RP11-439H9.1
|
RP11-439H9.1 |
| chr16_-_58718638 | 0.42 |
ENST00000562397.1
ENST00000564010.1 ENST00000570214.1 ENST00000563196.1 |
SLC38A7
|
solute carrier family 38, member 7 |
| chr7_-_38407770 | 0.41 |
ENST00000390348.2
|
TRGV1
|
T cell receptor gamma variable 1 (non-functional) |
| chr12_+_10460417 | 0.41 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chrX_-_100129320 | 0.40 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr6_-_111136513 | 0.39 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr4_-_130692631 | 0.39 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr10_+_97803151 | 0.39 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr3_-_114343768 | 0.39 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr21_-_34915123 | 0.36 |
ENST00000438059.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr2_+_191045562 | 0.35 |
ENST00000340623.4
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr7_+_157318477 | 0.35 |
ENST00000444154.1
|
AC006372.1
|
AC006372.1 |
| chr7_-_83824169 | 0.35 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr6_-_111804905 | 0.34 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr4_+_106631966 | 0.34 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr4_-_169401628 | 0.32 |
ENST00000514748.1
ENST00000512371.1 ENST00000260184.7 ENST00000505890.1 ENST00000511577.1 |
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
| chr2_-_202563414 | 0.32 |
ENST00000409474.3
ENST00000315506.7 ENST00000359962.5 |
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr17_-_39165366 | 0.31 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr1_-_161008697 | 0.31 |
ENST00000318289.10
ENST00000368023.3 ENST00000368024.1 ENST00000423014.2 |
TSTD1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr7_-_77427676 | 0.30 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr12_+_102514019 | 0.29 |
ENST00000537257.1
ENST00000358383.5 ENST00000392911.2 |
PARPBP
|
PARP1 binding protein |
| chrX_+_102024075 | 0.28 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr12_-_108955070 | 0.26 |
ENST00000228284.3
ENST00000546611.1 |
SART3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr1_-_151431647 | 0.25 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr14_+_24970800 | 0.25 |
ENST00000555109.1
|
RP11-80A15.1
|
Uncharacterized protein |
| chrY_+_14958970 | 0.25 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr3_-_108248169 | 0.25 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr11_-_65381643 | 0.23 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr12_-_108954933 | 0.23 |
ENST00000431469.2
ENST00000546815.1 |
SART3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr11_+_49050504 | 0.22 |
ENST00000332682.7
|
TRIM49B
|
tripartite motif containing 49B |
| chr2_-_166060382 | 0.21 |
ENST00000409101.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr4_-_76912070 | 0.20 |
ENST00000395711.4
ENST00000356260.5 |
SDAD1
|
SDA1 domain containing 1 |
| chr1_+_113010056 | 0.20 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr17_-_40337470 | 0.19 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr19_-_28957557 | 0.17 |
ENST00000585697.1
ENST00000591920.1 |
AC005307.3
|
AC005307.3 |
| chrX_+_119737806 | 0.16 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr2_-_202562774 | 0.15 |
ENST00000396886.3
ENST00000409143.1 |
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr3_+_193809263 | 0.15 |
ENST00000457815.1
|
RP11-135A1.3
|
RP11-135A1.3 |
| chr20_-_50419055 | 0.14 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr20_-_50418947 | 0.14 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr1_-_151431674 | 0.14 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chr2_-_166060571 | 0.14 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr5_+_67586465 | 0.13 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr6_-_26235206 | 0.11 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chrX_-_100129128 | 0.10 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr11_-_4414880 | 0.10 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr9_+_107456660 | 0.09 |
ENST00000318763.5
|
OR13D1
|
olfactory receptor, family 13, subfamily D, member 1 |
| chr13_+_49551020 | 0.08 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr2_-_202563351 | 0.07 |
ENST00000602867.1
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr22_-_28490123 | 0.07 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr9_+_103790991 | 0.06 |
ENST00000374874.3
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr1_+_171107241 | 0.06 |
ENST00000236166.3
|
FMO6P
|
flavin containing monooxygenase 6 pseudogene |
| chr9_-_118687405 | 0.06 |
ENST00000374014.3
|
LINC00474
|
long intergenic non-protein coding RNA 474 |
| chr20_-_50418972 | 0.05 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr12_-_30887948 | 0.05 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr9_-_86955598 | 0.04 |
ENST00000376238.4
|
SLC28A3
|
solute carrier family 28 (concentrative nucleoside transporter), member 3 |
| chr14_-_21109850 | 0.04 |
ENST00000320704.3
|
OR6S1
|
olfactory receptor, family 6, subfamily S, member 1 |
| chr2_-_166060552 | 0.03 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr3_+_77088989 | 0.03 |
ENST00000461745.1
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr6_+_25754927 | 0.03 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr19_-_58662139 | 0.03 |
ENST00000598312.1
|
ZNF329
|
zinc finger protein 329 |
| chr5_+_162864575 | 0.01 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr7_+_148287657 | 0.01 |
ENST00000307003.2
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr2_-_202562716 | 0.01 |
ENST00000428900.2
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU6F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:2000409 | astrocyte chemotaxis(GO:0035700) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) positive regulation of T cell extravasation(GO:2000409) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.4 | 1.8 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.3 | 0.9 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.6 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 2.2 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 0.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 1.7 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.1 | 0.5 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.4 | GO:0089709 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 1.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.6 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 1.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 1.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.8 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.9 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.6 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.6 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 1.4 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.5 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 1.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.8 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.2 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 2.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.6 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.9 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.9 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 2.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 1.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 2.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 1.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.4 | GO:0004644 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.1 | 0.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.4 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.5 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 2.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 1.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 3.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |