Project
Illumina Body Map 2
Navigation
Downloads
Results for PRDM1
Z-value: 1.66
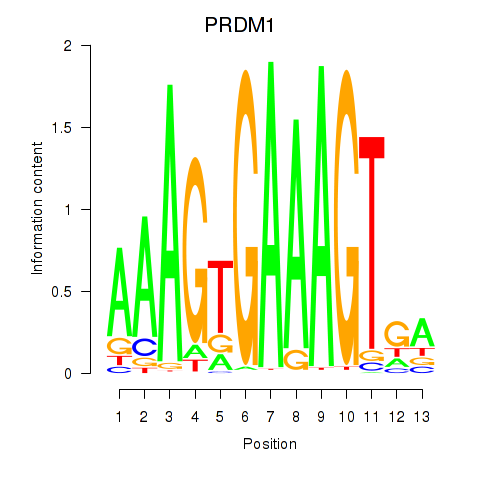
Transcription factors associated with PRDM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM1
|
ENSG00000057657.10 | PR/SET domain 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM1 | hg19_v2_chr6_+_106535455_106535637 | -0.12 | 5.2e-01 | Click! |
Activity profile of PRDM1 motif
Sorted Z-values of PRDM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_34710066 | 8.36 |
ENST00000378792.1
ENST00000259607.2 |
CCL21
|
chemokine (C-C motif) ligand 21 |
| chr10_-_44880491 | 7.52 |
ENST00000374426.2
ENST00000395795.4 ENST00000395794.2 ENST00000374429.2 ENST00000395793.3 ENST00000343575.6 |
CXCL12
|
chemokine (C-X-C motif) ligand 12 |
| chr6_-_112575912 | 7.19 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr6_-_112575687 | 6.96 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr6_-_112575758 | 6.78 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr4_-_100242549 | 6.40 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr2_+_239756671 | 6.17 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chr15_-_37393406 | 5.76 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr5_+_92919043 | 4.79 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr8_+_134203273 | 4.67 |
ENST00000250160.6
|
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr6_-_112575838 | 4.49 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr9_+_137533615 | 4.41 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chr3_+_8543533 | 4.35 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr6_-_29527702 | 4.33 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr10_-_131762105 | 4.15 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr11_+_46402583 | 4.05 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr3_-_168864427 | 3.95 |
ENST00000468789.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr21_-_40033618 | 3.79 |
ENST00000417133.2
ENST00000398910.1 ENST00000442448.1 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr3_+_8543561 | 3.72 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr8_+_134203303 | 3.70 |
ENST00000519433.1
ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr6_+_127439749 | 3.67 |
ENST00000356698.4
|
RSPO3
|
R-spondin 3 |
| chr11_+_46402482 | 3.66 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr3_+_8543393 | 3.65 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr9_+_18474098 | 3.55 |
ENST00000327883.7
ENST00000431052.2 ENST00000380570.4 |
ADAMTSL1
|
ADAMTS-like 1 |
| chr11_+_46402744 | 3.48 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr22_-_30642782 | 3.44 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr1_-_44497024 | 3.13 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr11_+_46402297 | 3.06 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr7_+_134464414 | 3.00 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr7_+_134464376 | 3.00 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr4_+_15376165 | 2.99 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr2_+_69240302 | 2.68 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr1_-_44497118 | 2.49 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr12_-_71003568 | 2.46 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr3_-_149375783 | 2.46 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr2_+_69240415 | 2.43 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chrX_+_114874727 | 2.39 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr6_+_118869452 | 2.33 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr8_+_97506033 | 2.27 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr1_+_212782012 | 2.25 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr1_+_199996733 | 2.19 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr8_-_54436491 | 2.19 |
ENST00000426023.1
|
RP11-400K9.4
|
RP11-400K9.4 |
| chr1_+_199996702 | 2.07 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr9_+_18474163 | 2.05 |
ENST00000380566.4
|
ADAMTSL1
|
ADAMTS-like 1 |
| chr7_+_15728003 | 2.05 |
ENST00000442176.1
|
AC005550.4
|
AC005550.4 |
| chr22_-_30642728 | 2.04 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr2_+_234526272 | 1.98 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr4_+_186990298 | 1.96 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr9_+_18474204 | 1.95 |
ENST00000276935.6
|
ADAMTSL1
|
ADAMTS-like 1 |
| chr13_+_102104980 | 1.92 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr19_-_43032532 | 1.86 |
ENST00000403461.1
ENST00000352591.5 ENST00000358394.3 ENST00000403444.3 ENST00000308072.4 ENST00000599389.1 ENST00000351134.3 ENST00000161559.6 |
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
| chr7_-_122526499 | 1.86 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr8_+_39770803 | 1.85 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr15_-_37392086 | 1.82 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr10_+_18429606 | 1.79 |
ENST00000324631.7
ENST00000352115.6 ENST00000377328.1 |
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr6_-_167275991 | 1.79 |
ENST00000510118.1
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr1_+_110453203 | 1.78 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr4_-_100140331 | 1.74 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr6_+_152011628 | 1.73 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr1_+_110453608 | 1.72 |
ENST00000369801.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chrX_-_154563889 | 1.71 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr1_+_110453109 | 1.70 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr2_-_36779411 | 1.64 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr2_+_234590556 | 1.61 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr15_+_71184931 | 1.60 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr6_-_105584560 | 1.58 |
ENST00000336775.5
|
BVES
|
blood vessel epicardial substance |
| chr10_+_18429671 | 1.58 |
ENST00000282343.8
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr1_+_110453514 | 1.58 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr20_+_6748311 | 1.57 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr9_+_133259836 | 1.50 |
ENST00000455439.2
|
HMCN2
|
hemicentin 2 |
| chr3_-_46000064 | 1.47 |
ENST00000433878.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr6_-_167276033 | 1.46 |
ENST00000503859.1
ENST00000506565.1 |
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr22_-_36556821 | 1.43 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr2_+_234545092 | 1.43 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr6_+_26156551 | 1.40 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr1_+_110453462 | 1.33 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr7_-_41742697 | 1.31 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr13_+_24144509 | 1.27 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr9_+_135457530 | 1.26 |
ENST00000263610.2
|
BARHL1
|
BarH-like homeobox 1 |
| chr10_-_35104185 | 1.25 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr11_+_71709938 | 1.23 |
ENST00000393705.4
ENST00000337131.5 ENST00000531053.1 ENST00000404792.1 |
IL18BP
|
interleukin 18 binding protein |
| chr3_-_46000146 | 1.20 |
ENST00000438446.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr3_-_64431058 | 1.14 |
ENST00000564377.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr16_-_88851618 | 1.12 |
ENST00000301015.9
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1 |
| chr15_+_71185148 | 1.11 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr14_-_69262947 | 1.11 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr19_+_24009879 | 1.08 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr19_+_34287174 | 1.07 |
ENST00000587559.1
ENST00000588637.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr13_-_33924755 | 1.04 |
ENST00000439831.1
ENST00000567873.1 |
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr12_-_57505121 | 0.99 |
ENST00000538913.2
ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr11_+_71710648 | 0.98 |
ENST00000260049.5
|
IL18BP
|
interleukin 18 binding protein |
| chr5_+_73109339 | 0.98 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr4_+_147145709 | 0.96 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr2_+_231191875 | 0.93 |
ENST00000444636.1
ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L
|
SP140 nuclear body protein-like |
| chr12_-_120763739 | 0.92 |
ENST00000549767.1
|
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr14_-_69262916 | 0.91 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr17_-_79818354 | 0.90 |
ENST00000576541.1
ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr6_-_105585022 | 0.90 |
ENST00000314641.5
|
BVES
|
blood vessel epicardial substance |
| chr11_+_64008525 | 0.88 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr5_-_66942617 | 0.87 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr20_-_57089934 | 0.86 |
ENST00000439429.1
ENST00000371149.3 |
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like |
| chr11_+_71710973 | 0.86 |
ENST00000393707.4
|
IL18BP
|
interleukin 18 binding protein |
| chr6_-_154831779 | 0.85 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr14_-_69262789 | 0.85 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr3_-_4793274 | 0.85 |
ENST00000414938.1
|
EGOT
|
eosinophil granule ontogeny transcript (non-protein coding) |
| chr17_+_41158742 | 0.84 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr13_+_24144796 | 0.84 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr2_+_234545148 | 0.83 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr16_+_20685815 | 0.82 |
ENST00000561584.1
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr2_+_231280908 | 0.81 |
ENST00000427101.2
ENST00000432979.1 |
SP100
|
SP100 nuclear antigen |
| chr11_+_64008443 | 0.81 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chrX_-_139015153 | 0.79 |
ENST00000370557.1
|
ATP11C
|
ATPase, class VI, type 11C |
| chr4_+_152020789 | 0.78 |
ENST00000512690.1
ENST00000508783.1 ENST00000512797.1 ENST00000507327.1 ENST00000515792.1 ENST00000506126.1 |
RPS3A
|
ribosomal protein S3A |
| chr1_+_2985726 | 0.77 |
ENST00000511072.1
ENST00000378398.3 ENST00000441472.2 ENST00000442529.2 |
PRDM16
|
PR domain containing 16 |
| chr1_+_164600184 | 0.76 |
ENST00000482110.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr14_-_69263043 | 0.74 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr8_-_79717163 | 0.72 |
ENST00000520269.1
|
IL7
|
interleukin 7 |
| chr6_+_130339710 | 0.71 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr4_+_146403912 | 0.71 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr7_+_143747461 | 0.70 |
ENST00000408906.2
|
OR2A5
|
olfactory receptor, family 2, subfamily A, member 5 |
| chr19_+_13106383 | 0.70 |
ENST00000397661.2
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr4_+_152020715 | 0.68 |
ENST00000274065.4
|
RPS3A
|
ribosomal protein S3A |
| chr4_+_152020736 | 0.68 |
ENST00000509736.1
ENST00000505243.1 ENST00000514682.1 ENST00000322686.6 ENST00000503002.1 |
RPS3A
|
ribosomal protein S3A |
| chr3_-_49851313 | 0.67 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr10_+_62538089 | 0.65 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr10_-_81203972 | 0.63 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr1_+_215747118 | 0.62 |
ENST00000448333.1
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr15_-_96607616 | 0.62 |
ENST00000558860.1
|
RP11-236L14.1
|
RP11-236L14.1 |
| chr2_+_231280954 | 0.62 |
ENST00000409824.1
ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100
|
SP100 nuclear antigen |
| chr1_+_2487631 | 0.60 |
ENST00000409119.1
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr2_-_202508169 | 0.59 |
ENST00000409883.2
|
TMEM237
|
transmembrane protein 237 |
| chr1_+_207262881 | 0.59 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr12_-_57504975 | 0.59 |
ENST00000553397.1
ENST00000556259.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr6_-_10412600 | 0.58 |
ENST00000379608.3
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr6_+_53659746 | 0.57 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr12_-_50561075 | 0.57 |
ENST00000422340.2
ENST00000317551.6 |
CERS5
|
ceramide synthase 5 |
| chr19_+_34287751 | 0.56 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr9_-_117568365 | 0.54 |
ENST00000374045.4
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr8_+_77593474 | 0.54 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr10_-_21661870 | 0.54 |
ENST00000433460.1
|
RP11-275N1.1
|
RP11-275N1.1 |
| chr2_-_70475586 | 0.53 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr10_+_62538248 | 0.51 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr3_+_119013185 | 0.51 |
ENST00000264245.4
|
ARHGAP31
|
Rho GTPase activating protein 31 |
| chr12_+_6561190 | 0.50 |
ENST00000544021.1
ENST00000266556.7 |
TAPBPL
|
TAP binding protein-like |
| chr17_-_26695013 | 0.50 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr4_-_146859787 | 0.50 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr8_+_9009202 | 0.47 |
ENST00000518496.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr8_-_9009079 | 0.47 |
ENST00000519699.1
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B |
| chr5_+_140430979 | 0.45 |
ENST00000306549.3
|
PCDHB1
|
protocadherin beta 1 |
| chr8_-_23540402 | 0.45 |
ENST00000523261.1
ENST00000380871.4 |
NKX3-1
|
NK3 homeobox 1 |
| chr21_+_22370717 | 0.43 |
ENST00000284894.7
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr1_-_205325994 | 0.42 |
ENST00000491471.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr7_-_111424506 | 0.42 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr7_+_95401851 | 0.42 |
ENST00000447467.2
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr7_-_47621229 | 0.41 |
ENST00000434451.1
|
TNS3
|
tensin 3 |
| chr6_-_9933500 | 0.40 |
ENST00000492169.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr3_+_69915385 | 0.40 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr17_-_26694979 | 0.39 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr5_+_86563636 | 0.38 |
ENST00000274376.6
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr7_-_86849025 | 0.37 |
ENST00000257637.3
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr17_+_7211656 | 0.37 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr14_-_21490958 | 0.36 |
ENST00000554104.1
|
NDRG2
|
NDRG family member 2 |
| chr1_-_205326161 | 0.36 |
ENST00000367156.3
ENST00000606887.1 ENST00000607173.1 |
KLHDC8A
|
kelch domain containing 8A |
| chr7_-_111424462 | 0.35 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr12_-_106641728 | 0.35 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr9_+_133454943 | 0.33 |
ENST00000319725.9
|
FUBP3
|
far upstream element (FUSE) binding protein 3 |
| chr2_-_64246206 | 0.32 |
ENST00000409558.4
ENST00000272322.4 |
VPS54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr8_-_56986768 | 0.32 |
ENST00000523936.1
|
RPS20
|
ribosomal protein S20 |
| chr17_+_6659354 | 0.32 |
ENST00000574907.1
|
XAF1
|
XIAP associated factor 1 |
| chr8_-_142012169 | 0.31 |
ENST00000517453.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr7_-_78400364 | 0.30 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr10_+_91589309 | 0.28 |
ENST00000448490.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr3_-_44552094 | 0.28 |
ENST00000436261.1
|
ZNF852
|
zinc finger protein 852 |
| chr6_-_138820624 | 0.28 |
ENST00000343505.5
|
NHSL1
|
NHS-like 1 |
| chr1_-_205326022 | 0.26 |
ENST00000367155.3
|
KLHDC8A
|
kelch domain containing 8A |
| chr1_+_2985760 | 0.26 |
ENST00000378391.2
ENST00000514189.1 ENST00000270722.5 |
PRDM16
|
PR domain containing 16 |
| chr5_+_76506706 | 0.24 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr6_-_32811771 | 0.24 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr5_-_93447333 | 0.24 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr18_-_47017956 | 0.23 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr1_+_199996859 | 0.23 |
ENST00000447034.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr17_-_57232525 | 0.22 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr4_+_142557717 | 0.22 |
ENST00000320650.4
ENST00000296545.7 |
IL15
|
interleukin 15 |
| chr10_+_91589261 | 0.21 |
ENST00000448963.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr1_-_111746966 | 0.19 |
ENST00000369752.5
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr15_-_56209306 | 0.19 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr1_-_161014731 | 0.19 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr22_+_39417118 | 0.18 |
ENST00000216099.8
|
APOBEC3D
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D |
| chr3_+_69915363 | 0.18 |
ENST00000451708.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr14_+_24605389 | 0.17 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr20_-_18774614 | 0.16 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr5_-_149380698 | 0.14 |
ENST00000296736.3
|
TIGD6
|
tigger transposable element derived 6 |
| chr6_+_57182400 | 0.14 |
ENST00000607273.1
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa) |
| chr11_-_96076334 | 0.14 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr4_-_140544386 | 0.13 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr6_-_121655552 | 0.12 |
ENST00000275159.6
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr11_+_128563652 | 0.12 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr9_+_84304628 | 0.12 |
ENST00000437181.1
|
RP11-154D17.1
|
RP11-154D17.1 |
| chr22_+_39436862 | 0.11 |
ENST00000381565.2
ENST00000452957.2 |
APOBEC3F
APOBEC3G
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G |
| chr21_+_17909594 | 0.11 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 2.4 | 14.2 | GO:0030421 | defecation(GO:0030421) |
| 1.9 | 5.6 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.7 | 5.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 1.6 | 8.1 | GO:1903971 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 1.5 | 7.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.5 | 4.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.1 | 5.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.8 | 2.5 | GO:0060921 | sinoatrial node development(GO:0003163) sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.8 | 2.3 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.7 | 2.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.7 | 3.6 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.6 | 1.9 | GO:0043317 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.5 | 3.8 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.5 | 7.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.5 | 5.8 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.5 | 1.6 | GO:0060129 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) regulation of calcium-independent cell-cell adhesion(GO:0051040) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.5 | 4.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.5 | 2.0 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.5 | 1.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.4 | 1.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.4 | 3.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.4 | 4.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.4 | 8.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.4 | 11.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 | 2.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 1.6 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.3 | 1.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 2.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 1.9 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 0.9 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.2 | 1.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.2 | 3.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 1.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.5 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.2 | 1.2 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.2 | 1.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.7 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 2.5 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.8 | GO:0090175 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.1 | 25.4 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.1 | 3.1 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.5 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.1 | 4.0 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.5 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.8 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 1.7 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 6.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 2.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.6 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.9 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.8 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 7.6 | GO:0008542 | visual learning(GO:0008542) pancreas development(GO:0031016) |
| 0.1 | 1.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.6 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 0.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.7 | GO:0031053 | osteoblast fate commitment(GO:0002051) primary miRNA processing(GO:0031053) |
| 0.0 | 0.3 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.4 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.3 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.2 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 1.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.6 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.9 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 5.1 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.7 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.6 | GO:0030111 | regulation of Wnt signaling pathway(GO:0030111) |
| 0.0 | 2.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.3 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 8.4 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 7.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.3 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.2 | 8.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.7 | 2.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.5 | 25.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.4 | 1.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 6.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 1.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 1.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 0.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 3.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.4 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 6.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 2.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 2.0 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 5.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 2.3 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.3 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 1.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 3.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 31.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 2.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 5.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 3.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 2.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 2.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 2.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 3.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 1.4 | 5.6 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 1.0 | 3.1 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 1.0 | 8.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.9 | 8.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.5 | 4.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 7.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.5 | 1.8 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.4 | 3.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 2.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 1.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.4 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.2 | 29.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 8.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 1.7 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 8.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 6.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 1.6 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 5.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 4.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 3.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.9 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.5 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 18.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 2.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.8 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 1.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 2.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 2.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 23.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 4.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 7.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 5.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 3.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 2.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 7.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 22.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 25.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 8.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 4.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 5.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 7.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 3.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 7.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 5.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 14.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 5.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 3.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 3.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 7.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 2.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 10.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 6.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 2.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |