Project
Illumina Body Map 2
Navigation
Downloads
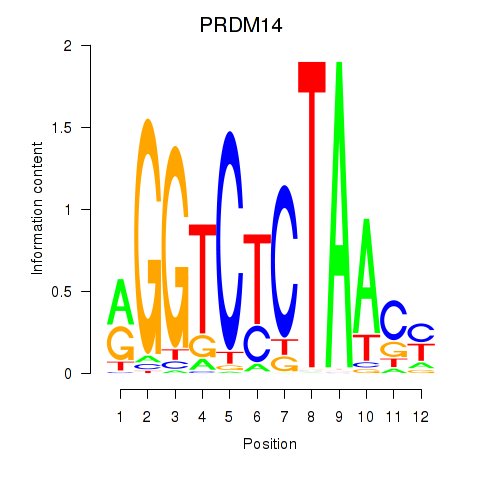
Results for PRDM14
Z-value: 0.36
Transcription factors associated with PRDM14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM14
|
ENSG00000147596.3 | PR/SET domain 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM14 | hg19_v2_chr8_-_70983506_70983562 | 0.03 | 8.8e-01 | Click! |
Activity profile of PRDM14 motif
Sorted Z-values of PRDM14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_41924511 | 1.10 |
ENST00000377203.4
ENST00000539718.1 ENST00000588884.1 ENST00000293396.8 ENST00000586233.1 |
CD300LG
|
CD300 molecule-like family member g |
| chr17_+_41924536 | 1.04 |
ENST00000317310.4
|
CD300LG
|
CD300 molecule-like family member g |
| chr17_-_46692287 | 0.93 |
ENST00000239144.4
|
HOXB8
|
homeobox B8 |
| chr17_-_46691990 | 0.58 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr6_-_46048116 | 0.50 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr4_-_10459009 | 0.49 |
ENST00000507515.1
|
ZNF518B
|
zinc finger protein 518B |
| chr2_-_192711968 | 0.48 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr1_+_36621174 | 0.48 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr1_+_36621529 | 0.43 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr3_-_134092561 | 0.32 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr17_-_19651668 | 0.31 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr17_+_36873677 | 0.31 |
ENST00000471200.1
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr11_+_64879317 | 0.31 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr14_-_54425475 | 0.29 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr4_-_10458982 | 0.29 |
ENST00000326756.3
|
ZNF518B
|
zinc finger protein 518B |
| chr1_+_156052354 | 0.26 |
ENST00000368301.2
|
LMNA
|
lamin A/C |
| chr2_-_228244013 | 0.26 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr5_+_32711419 | 0.25 |
ENST00000265074.8
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chrX_+_15525426 | 0.25 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr4_+_141677577 | 0.24 |
ENST00000609937.1
|
RP11-102N12.3
|
RP11-102N12.3 |
| chr6_-_109777128 | 0.23 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr2_-_145278475 | 0.23 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr17_-_19651654 | 0.23 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr17_-_17485731 | 0.21 |
ENST00000395783.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr19_+_3572758 | 0.19 |
ENST00000416526.1
|
HMG20B
|
high mobility group 20B |
| chr17_-_2117600 | 0.19 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr8_+_101349823 | 0.19 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr1_-_33642151 | 0.19 |
ENST00000543586.1
|
TRIM62
|
tripartite motif containing 62 |
| chr17_-_46692457 | 0.18 |
ENST00000468443.1
|
HOXB8
|
homeobox B8 |
| chr5_+_32710736 | 0.18 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr19_-_5624057 | 0.18 |
ENST00000590262.1
|
SAFB2
|
scaffold attachment factor B2 |
| chr3_+_155838337 | 0.17 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr2_-_145277882 | 0.17 |
ENST00000465070.1
ENST00000444559.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_155006300 | 0.17 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr17_-_19651598 | 0.16 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr7_+_150211918 | 0.15 |
ENST00000313543.4
|
GIMAP7
|
GTPase, IMAP family member 7 |
| chr19_-_49314269 | 0.14 |
ENST00000545387.2
ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr12_-_76742183 | 0.14 |
ENST00000393262.3
|
BBS10
|
Bardet-Biedl syndrome 10 |
| chr5_-_135528822 | 0.13 |
ENST00000607574.1
|
AC009014.3
|
AC009014.3 |
| chr9_+_87283430 | 0.13 |
ENST00000376214.1
ENST00000376213.1 |
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr3_-_143567262 | 0.12 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr17_-_73775839 | 0.12 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr2_+_113033164 | 0.12 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr11_-_62439727 | 0.11 |
ENST00000528862.1
|
C11orf48
|
chromosome 11 open reading frame 48 |
| chr19_-_49314169 | 0.10 |
ENST00000597011.1
ENST00000601681.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr4_+_4861385 | 0.10 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr20_-_48770174 | 0.09 |
ENST00000341698.2
|
TMEM189-UBE2V1
|
TMEM189-UBE2V1 readthrough |
| chr11_-_105892937 | 0.09 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr6_+_43140095 | 0.09 |
ENST00000457278.2
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor) |
| chr3_+_16926441 | 0.09 |
ENST00000418129.2
ENST00000396755.2 |
PLCL2
|
phospholipase C-like 2 |
| chr7_-_140714739 | 0.09 |
ENST00000467334.1
ENST00000324787.5 |
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr14_+_89290965 | 0.09 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr3_+_37035289 | 0.08 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr9_+_132099158 | 0.08 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr7_-_140714430 | 0.08 |
ENST00000393008.3
|
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr6_+_133561725 | 0.08 |
ENST00000452339.2
|
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr11_-_46615498 | 0.08 |
ENST00000533727.1
ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1
|
autophagy/beclin-1 regulator 1 |
| chr12_-_124456598 | 0.07 |
ENST00000539761.1
ENST00000539551.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chr5_-_108063949 | 0.07 |
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023 |
| chr8_+_110346546 | 0.06 |
ENST00000521662.1
ENST00000521688.1 ENST00000520147.1 |
ENY2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr1_-_31845914 | 0.06 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr17_+_75450075 | 0.06 |
ENST00000592951.1
|
SEPT9
|
septin 9 |
| chrX_+_41193407 | 0.06 |
ENST00000457138.2
ENST00000441189.2 |
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr19_+_39109735 | 0.06 |
ENST00000593149.1
ENST00000248342.4 ENST00000538434.1 ENST00000588934.1 ENST00000545173.2 ENST00000589307.1 ENST00000586513.1 ENST00000591409.1 ENST00000592558.1 |
EIF3K
|
eukaryotic translation initiation factor 3, subunit K |
| chr5_-_55777586 | 0.05 |
ENST00000506836.1
|
CTC-236F12.4
|
Uncharacterized protein |
| chr2_-_208030295 | 0.05 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_-_47517302 | 0.05 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr2_-_208030647 | 0.05 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_+_10289707 | 0.05 |
ENST00000287652.4
|
TATDN2
|
TatD DNase domain containing 2 |
| chr12_-_56326402 | 0.04 |
ENST00000547925.1
|
WIBG
|
within bgcn homolog (Drosophila) |
| chr8_-_102216925 | 0.04 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr3_+_37035263 | 0.04 |
ENST00000458205.2
ENST00000539477.1 |
MLH1
|
mutL homolog 1 |
| chr12_-_49351303 | 0.04 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr12_-_49351228 | 0.04 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr17_+_75450146 | 0.04 |
ENST00000585929.1
ENST00000590917.1 |
SEPT9
|
septin 9 |
| chr9_-_125148928 | 0.03 |
ENST00000600713.1
|
AL162424.1
|
Uncharacterized protein |
| chr1_+_153963227 | 0.03 |
ENST00000368567.4
ENST00000392558.4 |
RPS27
|
ribosomal protein S27 |
| chr7_+_39989611 | 0.03 |
ENST00000181839.4
|
CDK13
|
cyclin-dependent kinase 13 |
| chr17_+_75450099 | 0.03 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr7_-_30066233 | 0.03 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr1_+_45274154 | 0.03 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr17_+_47653178 | 0.03 |
ENST00000328741.5
|
NXPH3
|
neurexophilin 3 |
| chr13_-_34250861 | 0.03 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr15_+_45694567 | 0.03 |
ENST00000559860.1
|
SPATA5L1
|
spermatogenesis associated 5-like 1 |
| chr14_-_21492251 | 0.02 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr19_-_53757941 | 0.02 |
ENST00000594517.1
ENST00000601413.1 ENST00000594681.1 |
ZNF677
|
zinc finger protein 677 |
| chr2_-_37193606 | 0.02 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr1_-_42384343 | 0.01 |
ENST00000372584.1
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr15_+_45694523 | 0.01 |
ENST00000305560.6
|
SPATA5L1
|
spermatogenesis associated 5-like 1 |
| chr2_-_208030886 | 0.01 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_+_65730385 | 0.01 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr17_+_39994032 | 0.01 |
ENST00000293303.4
ENST00000438813.1 |
KLHL10
|
kelch-like family member 10 |
| chr19_-_6279932 | 0.00 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.2 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.3 | GO:0048391 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 1.7 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0060532 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.2 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 2.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |