Project
Illumina Body Map 2
Navigation
Downloads
Results for RFX3_RFX2
Z-value: 7.20
Transcription factors associated with RFX3_RFX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
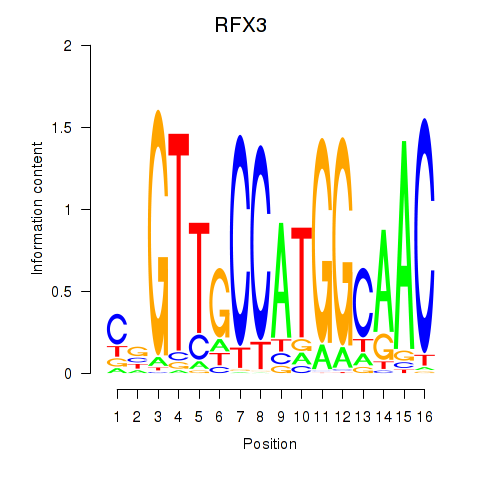
RFX3
|
ENSG00000080298.11 | regulatory factor X3 |
|
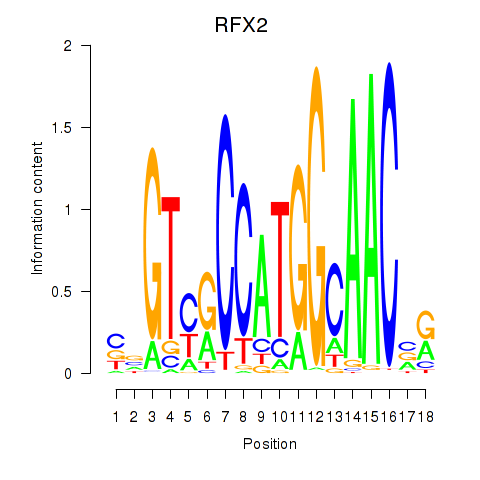
RFX2
|
ENSG00000087903.8 | regulatory factor X2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX2 | hg19_v2_chr19_-_6199521_6199546 | 0.74 | 1.4e-06 | Click! |
| RFX3 | hg19_v2_chr9_-_3489428_3489437 | 0.60 | 2.6e-04 | Click! |
Activity profile of RFX3_RFX2 motif
Sorted Z-values of RFX3_RFX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_109656439 | 37.05 |
ENST00000369949.4
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr21_-_43916296 | 30.70 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr21_-_43916433 | 29.91 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr1_-_169396666 | 29.38 |
ENST00000456107.1
ENST00000367805.3 |
CCDC181
|
coiled-coil domain containing 181 |
| chr1_-_169396646 | 28.04 |
ENST00000367806.3
|
CCDC181
|
coiled-coil domain containing 181 |
| chr1_-_159869912 | 22.71 |
ENST00000368099.4
|
CCDC19
|
coiled-coil domain containing 19 |
| chr5_+_34839260 | 22.51 |
ENST00000505624.1
|
TTC23L
|
tetratricopeptide repeat domain 23-like |
| chr3_-_167098059 | 21.68 |
ENST00000392764.1
ENST00000474464.1 ENST00000392766.2 ENST00000485651.1 |
ZBBX
|
zinc finger, B-box domain containing |
| chr17_+_9479944 | 21.66 |
ENST00000396219.3
ENST00000352665.5 |
WDR16
|
WD repeat domain 16 |
| chr16_+_84178874 | 20.30 |
ENST00000378553.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr17_-_74137374 | 20.23 |
ENST00000322957.6
|
FOXJ1
|
forkhead box J1 |
| chr10_+_63422695 | 20.15 |
ENST00000330194.2
ENST00000389639.3 |
C10orf107
|
chromosome 10 open reading frame 107 |
| chr16_+_84178850 | 19.65 |
ENST00000334315.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr1_-_173638976 | 19.39 |
ENST00000333279.2
|
ANKRD45
|
ankyrin repeat domain 45 |
| chr9_+_124922171 | 19.34 |
ENST00000373764.3
ENST00000536616.1 |
MORN5
|
MORN repeat containing 5 |
| chr11_-_75379479 | 19.27 |
ENST00000434603.2
|
MAP6
|
microtubule-associated protein 6 |
| chr2_+_228736321 | 19.04 |
ENST00000309931.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr15_+_71184931 | 18.40 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr20_-_3762087 | 18.35 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr1_-_118727781 | 18.31 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
| chr11_-_75379612 | 18.09 |
ENST00000526740.1
|
MAP6
|
microtubule-associated protein 6 |
| chr17_-_6735035 | 17.44 |
ENST00000338694.2
|
TEKT1
|
tektin 1 |
| chr8_-_49647791 | 17.43 |
ENST00000262103.3
ENST00000523092.1 ENST00000433756.1 |
EFCAB1
|
EF-hand calcium binding domain 1 |
| chr12_+_48577366 | 17.28 |
ENST00000316554.3
|
C12orf68
|
chromosome 12 open reading frame 68 |
| chr19_-_57352064 | 17.27 |
ENST00000326441.9
ENST00000593695.1 ENST00000599577.1 ENST00000594389.1 ENST00000423103.2 ENST00000598410.1 ENST00000593711.1 ENST00000391708.3 ENST00000221722.5 ENST00000599935.1 |
PEG3
ZIM2
|
paternally expressed 3 zinc finger, imprinted 2 |
| chr12_+_85430110 | 17.27 |
ENST00000393212.3
ENST00000393217.2 |
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1 |
| chr11_+_94245617 | 17.11 |
ENST00000542198.1
|
RP11-867G2.2
|
long intergenic non-protein coding RNA 1171 |
| chrX_-_48693955 | 16.86 |
ENST00000218230.5
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr4_-_177116772 | 16.83 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr17_-_6735012 | 16.74 |
ENST00000535086.1
|
TEKT1
|
tektin 1 |
| chr6_-_165723088 | 16.52 |
ENST00000230301.8
|
C6orf118
|
chromosome 6 open reading frame 118 |
| chr15_+_43803143 | 16.22 |
ENST00000382031.1
|
MAP1A
|
microtubule-associated protein 1A |
| chr3_-_50383096 | 16.16 |
ENST00000442887.1
ENST00000360165.3 |
ZMYND10
|
zinc finger, MYND-type containing 10 |
| chr1_+_151682909 | 15.89 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr16_+_57728701 | 15.66 |
ENST00000569375.1
ENST00000360716.3 ENST00000569167.1 ENST00000394337.4 ENST00000563126.1 ENST00000336825.8 |
CCDC135
|
coiled-coil domain containing 135 |
| chr1_-_154178803 | 15.57 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr13_+_37005967 | 15.44 |
ENST00000440264.1
ENST00000449823.1 |
CCNA1
|
cyclin A1 |
| chr4_-_153700864 | 15.35 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chr15_+_71185148 | 15.29 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr9_+_135285579 | 15.04 |
ENST00000343036.2
ENST00000393216.2 |
C9orf171
|
chromosome 9 open reading frame 171 |
| chr4_-_76555657 | 15.00 |
ENST00000307465.4
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr12_+_106994905 | 14.35 |
ENST00000357881.4
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr1_+_151693984 | 14.25 |
ENST00000479191.1
|
RIIAD1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 |
| chr2_-_130902567 | 14.21 |
ENST00000457413.1
ENST00000392984.3 ENST00000409128.1 ENST00000441670.1 ENST00000409943.3 ENST00000409234.3 ENST00000310463.6 |
CCDC74B
|
coiled-coil domain containing 74B |
| chr2_+_120302041 | 14.17 |
ENST00000442513.3
ENST00000413369.3 |
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr3_+_126113734 | 14.12 |
ENST00000352312.1
ENST00000393425.1 |
CCDC37
|
coiled-coil domain containing 37 |
| chr5_-_149669192 | 13.91 |
ENST00000398376.3
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr8_+_144798429 | 13.53 |
ENST00000338033.4
ENST00000395107.4 ENST00000395108.2 |
MAPK15
|
mitogen-activated protein kinase 15 |
| chr6_-_35109080 | 13.48 |
ENST00000486638.1
ENST00000505400.1 ENST00000412155.2 ENST00000373979.2 ENST00000507706.1 ENST00000444780.2 ENST00000492680.2 |
TCP11
|
t-complex 11, testis-specific |
| chr2_+_120301997 | 13.42 |
ENST00000602047.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr1_+_43637996 | 13.39 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr2_+_79740118 | 13.05 |
ENST00000496558.1
ENST00000451966.1 |
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr19_+_54385439 | 13.03 |
ENST00000536044.1
ENST00000540413.1 ENST00000263431.3 ENST00000419486.1 |
PRKCG
|
protein kinase C, gamma |
| chr12_+_51818555 | 12.96 |
ENST00000453097.2
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr10_+_127585093 | 12.87 |
ENST00000368695.1
ENST00000368693.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr19_-_46974741 | 12.86 |
ENST00000313683.10
ENST00000602246.1 |
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr11_-_8615488 | 12.79 |
ENST00000315204.1
ENST00000396672.1 ENST00000396673.1 |
STK33
|
serine/threonine kinase 33 |
| chr6_-_43478239 | 12.68 |
ENST00000372441.1
|
LRRC73
|
leucine rich repeat containing 73 |
| chr4_-_87515202 | 12.58 |
ENST00000502302.1
ENST00000513186.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr8_-_101118185 | 12.38 |
ENST00000523437.1
|
RGS22
|
regulator of G-protein signaling 22 |
| chr18_-_47792851 | 12.34 |
ENST00000398545.4
|
CCDC11
|
coiled-coil domain containing 11 |
| chr7_+_48075108 | 12.17 |
ENST00000420324.1
ENST00000435376.1 ENST00000430738.1 ENST00000348904.3 ENST00000539619.1 |
C7orf57
|
chromosome 7 open reading frame 57 |
| chr11_-_111944704 | 12.15 |
ENST00000532211.1
|
PIH1D2
|
PIH1 domain containing 2 |
| chr13_+_31506818 | 12.09 |
ENST00000380473.3
|
TEX26
|
testis expressed 26 |
| chr11_-_8615687 | 12.01 |
ENST00000534493.1
ENST00000422559.2 |
STK33
|
serine/threonine kinase 33 |
| chr15_+_82555125 | 12.01 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr9_-_135754164 | 12.01 |
ENST00000298545.3
|
AK8
|
adenylate kinase 8 |
| chr5_-_54529415 | 11.93 |
ENST00000282572.4
|
CCNO
|
cyclin O |
| chr1_-_6240183 | 11.90 |
ENST00000262450.3
ENST00000378021.1 |
CHD5
|
chromodomain helicase DNA binding protein 5 |
| chr2_+_27071045 | 11.88 |
ENST00000401478.1
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr1_+_42928945 | 11.86 |
ENST00000428554.2
|
CCDC30
|
coiled-coil domain containing 30 |
| chr1_-_183622442 | 11.85 |
ENST00000308641.4
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr22_-_39052300 | 11.84 |
ENST00000355830.6
|
FAM227A
|
family with sequence similarity 227, member A |
| chr16_+_67840986 | 11.76 |
ENST00000561639.1
ENST00000567852.1 ENST00000565148.1 ENST00000388833.3 ENST00000561654.1 ENST00000431934.2 |
TSNAXIP1
|
translin-associated factor X interacting protein 1 |
| chr1_+_161068179 | 11.73 |
ENST00000368011.4
ENST00000392192.2 |
KLHDC9
|
kelch domain containing 9 |
| chr19_-_46974664 | 11.70 |
ENST00000438932.2
|
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr15_+_82555169 | 11.64 |
ENST00000565432.1
ENST00000427381.2 |
FAM154B
|
family with sequence similarity 154, member B |
| chr3_-_118753716 | 11.56 |
ENST00000393775.2
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr2_-_233792837 | 11.53 |
ENST00000373552.4
ENST00000409079.1 |
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr1_+_38022572 | 11.47 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr17_+_9479971 | 11.44 |
ENST00000576499.1
|
WDR16
|
WD repeat domain 16 |
| chr5_-_114505624 | 11.35 |
ENST00000513154.1
|
TRIM36
|
tripartite motif containing 36 |
| chr11_-_8615720 | 11.32 |
ENST00000358872.3
ENST00000454443.2 |
STK33
|
serine/threonine kinase 33 |
| chr17_-_7108436 | 11.28 |
ENST00000493294.1
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr16_+_4784458 | 11.25 |
ENST00000590191.1
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr6_-_109761707 | 11.20 |
ENST00000520723.1
ENST00000518648.1 ENST00000417394.2 |
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr11_-_111944895 | 11.11 |
ENST00000431456.1
ENST00000280350.4 ENST00000530641.1 |
PIH1D2
|
PIH1 domain containing 2 |
| chr3_-_118753626 | 11.06 |
ENST00000489689.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr5_-_11589131 | 10.99 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr17_+_56833184 | 10.98 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr20_+_20033158 | 10.98 |
ENST00000340348.6
ENST00000377309.2 ENST00000389656.3 ENST00000377306.1 ENST00000245957.5 ENST00000377303.2 ENST00000475466.1 |
C20orf26
|
chromosome 20 open reading frame 26 |
| chr5_+_157098534 | 10.97 |
ENST00000409999.3
|
C5orf52
|
chromosome 5 open reading frame 52 |
| chrX_+_117957741 | 10.96 |
ENST00000310164.2
|
ZCCHC12
|
zinc finger, CCHC domain containing 12 |
| chr6_-_35109145 | 10.95 |
ENST00000373974.4
ENST00000244645.3 |
TCP11
|
t-complex 11, testis-specific |
| chr16_+_67840668 | 10.84 |
ENST00000415766.3
|
TSNAXIP1
|
translin-associated factor X interacting protein 1 |
| chr6_-_19804973 | 10.66 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr1_+_38022513 | 10.66 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr10_-_25305011 | 10.60 |
ENST00000331161.4
ENST00000376363.1 |
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr10_+_124030819 | 10.55 |
ENST00000260723.4
ENST00000368994.2 |
BTBD16
|
BTB (POZ) domain containing 16 |
| chr8_-_133687813 | 10.53 |
ENST00000250173.1
ENST00000519595.1 |
LRRC6
|
leucine rich repeat containing 6 |
| chr11_+_111385497 | 10.50 |
ENST00000375618.4
ENST00000529167.1 ENST00000332814.6 |
C11orf88
|
chromosome 11 open reading frame 88 |
| chr2_+_132285406 | 10.48 |
ENST00000295171.6
ENST00000409856.3 |
CCDC74A
|
coiled-coil domain containing 74A |
| chr2_-_2334888 | 10.39 |
ENST00000428368.2
ENST00000399161.2 |
MYT1L
|
myelin transcription factor 1-like |
| chr8_-_101118210 | 10.30 |
ENST00000360863.6
|
RGS22
|
regulator of G-protein signaling 22 |
| chr17_+_11501748 | 10.30 |
ENST00000262442.4
ENST00000579828.1 |
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr10_-_104179682 | 10.28 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr5_-_114505591 | 10.20 |
ENST00000508894.1
|
TRIM36
|
tripartite motif containing 36 |
| chr1_-_21978312 | 10.10 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr5_-_11588907 | 10.05 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr16_-_71264558 | 10.04 |
ENST00000448089.2
ENST00000393550.2 ENST00000448691.1 ENST00000393567.2 ENST00000321489.5 ENST00000539973.1 ENST00000288168.10 ENST00000545267.1 ENST00000541601.1 ENST00000538248.1 |
HYDIN
|
HYDIN, axonemal central pair apparatus protein |
| chr13_-_25745857 | 9.99 |
ENST00000381853.3
|
AMER2
|
APC membrane recruitment protein 2 |
| chr19_-_51289436 | 9.90 |
ENST00000562076.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr1_+_20512568 | 9.86 |
ENST00000375099.3
|
UBXN10
|
UBX domain protein 10 |
| chr3_-_118753566 | 9.83 |
ENST00000491903.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr16_+_27078219 | 9.69 |
ENST00000418886.1
|
C16orf82
|
chromosome 16 open reading frame 82 |
| chr20_+_30467600 | 9.64 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr17_-_60885645 | 9.59 |
ENST00000544856.2
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr19_+_58545434 | 9.58 |
ENST00000282326.1
ENST00000601162.1 |
ZSCAN1
|
zinc finger and SCAN domain containing 1 |
| chr17_-_60885700 | 9.57 |
ENST00000583600.1
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr17_-_70417365 | 9.56 |
ENST00000580948.1
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr11_-_66496430 | 9.55 |
ENST00000533211.1
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2 |
| chr1_+_160051319 | 9.49 |
ENST00000368088.3
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr2_+_27070964 | 9.45 |
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr3_+_119421849 | 9.44 |
ENST00000273390.5
ENST00000463700.1 |
MAATS1
|
MYCBP-associated, testis expressed 1 |
| chrX_+_85969626 | 9.39 |
ENST00000484479.1
|
DACH2
|
dachshund homolog 2 (Drosophila) |
| chr10_+_134973905 | 9.35 |
ENST00000304613.3
ENST00000368572.2 |
KNDC1
|
kinase non-catalytic C-lobe domain (KIND) containing 1 |
| chr4_-_87281196 | 9.32 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr1_-_40157345 | 9.30 |
ENST00000372844.3
|
HPCAL4
|
hippocalcin like 4 |
| chr6_+_116937636 | 9.28 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr6_-_24646249 | 9.24 |
ENST00000430948.2
ENST00000537886.1 ENST00000535378.1 ENST00000378214.3 |
KIAA0319
|
KIAA0319 |
| chr7_-_45128472 | 9.18 |
ENST00000490531.2
|
NACAD
|
NAC alpha domain containing |
| chr19_-_55690758 | 9.15 |
ENST00000590851.1
|
SYT5
|
synaptotagmin V |
| chr5_-_132073111 | 9.14 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr8_-_133687778 | 9.14 |
ENST00000518642.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr3_-_47324008 | 9.14 |
ENST00000425853.1
|
KIF9
|
kinesin family member 9 |
| chr11_-_8615507 | 9.13 |
ENST00000431279.2
ENST00000418597.1 |
STK33
|
serine/threonine kinase 33 |
| chr4_-_87281224 | 9.12 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr1_-_50489547 | 9.09 |
ENST00000371836.1
ENST00000371839.1 ENST00000371838.1 |
AGBL4
|
ATP/GTP binding protein-like 4 |
| chr21_+_18885430 | 9.09 |
ENST00000356275.6
ENST00000400165.1 ENST00000400169.1 ENST00000306618.10 |
CXADR
|
coxsackie virus and adenovirus receptor |
| chr2_-_187713891 | 9.02 |
ENST00000295131.2
|
ZSWIM2
|
zinc finger, SWIM-type containing 2 |
| chr17_-_60885659 | 9.02 |
ENST00000311269.5
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr16_-_30064244 | 8.97 |
ENST00000571269.1
ENST00000561666.1 |
FAM57B
|
family with sequence similarity 57, member B |
| chr3_-_27410847 | 8.90 |
ENST00000429845.2
ENST00000341435.5 ENST00000435750.1 |
NEK10
|
NIMA-related kinase 10 |
| chr22_+_30752963 | 8.87 |
ENST00000445005.1
ENST00000430839.1 |
CCDC157
|
coiled-coil domain containing 157 |
| chr21_-_40817645 | 8.79 |
ENST00000438404.1
ENST00000358268.2 ENST00000411566.1 ENST00000451131.1 ENST00000418018.1 ENST00000415863.1 ENST00000426783.1 ENST00000288350.3 ENST00000485895.2 ENST00000448288.2 ENST00000456017.1 ENST00000434281.1 |
LCA5L
|
Leber congenital amaurosis 5-like |
| chr1_-_220101944 | 8.73 |
ENST00000366926.3
ENST00000536992.1 |
SLC30A10
|
solute carrier family 30, member 10 |
| chr8_+_85618155 | 8.72 |
ENST00000523850.1
ENST00000521376.1 |
RALYL
|
RALY RNA binding protein-like |
| chr13_+_37006398 | 8.72 |
ENST00000418263.1
|
CCNA1
|
cyclin A1 |
| chr1_-_67390474 | 8.70 |
ENST00000371023.3
ENST00000371022.3 ENST00000371026.3 ENST00000431318.1 |
WDR78
|
WD repeat domain 78 |
| chr9_-_35812140 | 8.69 |
ENST00000497810.1
ENST00000396638.2 ENST00000484764.1 |
SPAG8
|
sperm associated antigen 8 |
| chr16_-_30022735 | 8.55 |
ENST00000564944.1
|
DOC2A
|
double C2-like domains, alpha |
| chr6_-_32119676 | 8.55 |
ENST00000211413.5
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chrX_-_152160740 | 8.54 |
ENST00000361887.5
ENST00000439251.1 ENST00000452693.1 |
PNMA5
|
paraneoplastic Ma antigen family member 5 |
| chr2_+_202937972 | 8.50 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr5_-_110074603 | 8.48 |
ENST00000515278.2
|
TMEM232
|
transmembrane protein 232 |
| chr13_+_37006421 | 8.42 |
ENST00000255465.4
|
CCNA1
|
cyclin A1 |
| chr1_+_33546714 | 8.38 |
ENST00000294517.6
ENST00000358680.3 ENST00000373443.3 ENST00000398167.1 |
ADC
|
arginine decarboxylase |
| chr1_+_109656719 | 8.37 |
ENST00000457623.2
ENST00000529753.1 |
KIAA1324
|
KIAA1324 |
| chr9_+_135285430 | 8.35 |
ENST00000393215.3
|
C9orf171
|
chromosome 9 open reading frame 171 |
| chr6_-_32119877 | 8.31 |
ENST00000375152.2
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr2_+_159651821 | 8.31 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr4_-_156297949 | 8.28 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr21_+_18885318 | 8.13 |
ENST00000400166.1
|
CXADR
|
coxsackie virus and adenovirus receptor |
| chr16_-_30773372 | 8.10 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr11_-_66360548 | 8.05 |
ENST00000333861.3
|
CCDC87
|
coiled-coil domain containing 87 |
| chr11_+_134201768 | 8.00 |
ENST00000535456.2
ENST00000339772.7 |
GLB1L2
|
galactosidase, beta 1-like 2 |
| chr3_-_149688896 | 8.00 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr11_-_119252359 | 7.99 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr7_-_102715172 | 7.99 |
ENST00000456695.1
ENST00000455112.2 ENST00000440067.1 |
FBXL13
|
F-box and leucine-rich repeat protein 13 |
| chrX_+_119029800 | 7.97 |
ENST00000371431.3
ENST00000371423.2 ENST00000371425.4 ENST00000394594.2 |
AKAP14
|
A kinase (PRKA) anchor protein 14 |
| chr2_+_232063260 | 7.97 |
ENST00000349938.4
|
ARMC9
|
armadillo repeat containing 9 |
| chr10_-_135150367 | 7.95 |
ENST00000368555.3
ENST00000252939.4 ENST00000368558.1 ENST00000368556.2 |
CALY
|
calcyon neuron-specific vesicular protein |
| chr7_+_129142320 | 7.94 |
ENST00000462322.2
|
SMKR1
|
small lysine-rich protein 1 |
| chr19_-_7990991 | 7.88 |
ENST00000318978.4
|
CTXN1
|
cortexin 1 |
| chr2_+_120302055 | 7.88 |
ENST00000598644.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr15_+_67547113 | 7.87 |
ENST00000512104.1
ENST00000358767.3 ENST00000546225.1 |
IQCH
|
IQ motif containing H |
| chr7_+_73275483 | 7.85 |
ENST00000320531.2
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28 |
| chr1_+_217804661 | 7.85 |
ENST00000366933.4
|
SPATA17
|
spermatogenesis associated 17 |
| chr5_-_132073210 | 7.82 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr19_+_58545369 | 7.76 |
ENST00000391700.1
|
ZSCAN1
|
zinc finger and SCAN domain containing 1 |
| chr11_+_101918153 | 7.76 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr16_+_29823427 | 7.72 |
ENST00000358758.7
ENST00000567659.1 ENST00000572820.1 |
PRRT2
|
proline-rich transmembrane protein 2 |
| chr12_+_122356488 | 7.71 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr7_+_133812052 | 7.70 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr1_+_109656579 | 7.70 |
ENST00000526264.1
ENST00000369939.3 |
KIAA1324
|
KIAA1324 |
| chr1_+_111889212 | 7.70 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr7_-_102715263 | 7.69 |
ENST00000379305.3
|
FBXL13
|
F-box and leucine-rich repeat protein 13 |
| chr4_+_87515454 | 7.65 |
ENST00000427191.2
ENST00000436978.1 ENST00000502971.1 |
PTPN13
|
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr8_-_37797621 | 7.60 |
ENST00000524298.1
ENST00000307599.4 |
GOT1L1
|
glutamic-oxaloacetic transaminase 1-like 1 |
| chr11_-_31391276 | 7.58 |
ENST00000452803.1
|
DCDC1
|
doublecortin domain containing 1 |
| chr4_+_40751914 | 7.57 |
ENST00000381782.2
ENST00000316607.5 |
NSUN7
|
NOP2/Sun domain family, member 7 |
| chr14_+_60386853 | 7.54 |
ENST00000570145.1
|
LRRC9
|
leucine rich repeat containing 9 |
| chr10_+_103348031 | 7.54 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr19_-_55677920 | 7.50 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr10_+_22634384 | 7.44 |
ENST00000376624.3
ENST00000376603.2 ENST00000376601.1 ENST00000538630.1 ENST00000456231.2 ENST00000313311.6 ENST00000435326.1 |
SPAG6
|
sperm associated antigen 6 |
| chr1_-_36916011 | 7.40 |
ENST00000356637.5
ENST00000354267.3 ENST00000235532.5 |
OSCP1
|
organic solute carrier partner 1 |
| chr11_-_119252425 | 7.40 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr15_-_71184724 | 7.37 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr1_+_15573757 | 7.37 |
ENST00000358897.4
ENST00000417793.1 ENST00000375999.3 ENST00000433640.2 |
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr3_-_118753792 | 7.33 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX3_RFX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 24.4 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 6.7 | 20.2 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 5.7 | 28.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 4.7 | 41.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 4.0 | 11.9 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 3.4 | 10.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 2.9 | 17.2 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 2.7 | 31.9 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 2.6 | 23.6 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 2.6 | 13.0 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 2.6 | 117.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 2.5 | 7.6 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 2.5 | 41.8 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 2.2 | 31.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 2.2 | 6.5 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 2.1 | 50.9 | GO:0003341 | cilium movement(GO:0003341) |
| 2.1 | 24.8 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 1.9 | 7.8 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.6 | 4.8 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 1.6 | 7.9 | GO:0098907 | protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 1.4 | 17.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 1.4 | 6.9 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.3 | 11.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 1.3 | 14.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 1.3 | 11.3 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 1.2 | 17.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 1.1 | 9.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 1.1 | 8.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.1 | 6.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.1 | 15.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 1.0 | 4.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 1.0 | 19.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 1.0 | 20.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 1.0 | 3.0 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 1.0 | 14.7 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.0 | 27.8 | GO:1904385 | cellular response to angiotensin(GO:1904385) |
| 0.9 | 9.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.9 | 5.5 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.9 | 11.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.8 | 9.2 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.8 | 11.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.8 | 32.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.8 | 12.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.8 | 4.7 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.8 | 1.6 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.8 | 4.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.7 | 3.7 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.7 | 9.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.7 | 28.3 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.7 | 2.1 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.7 | 14.6 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.6 | 5.8 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.6 | 4.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.6 | 8.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.6 | 31.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.6 | 5.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.5 | 4.2 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.5 | 2.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.5 | 3.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.5 | 15.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.5 | 2.0 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.5 | 1.9 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.5 | 16.9 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.5 | 5.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.5 | 5.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.5 | 8.2 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.4 | 3.5 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.4 | 9.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.4 | 1.6 | GO:0060166 | olfactory pit development(GO:0060166) Harderian gland development(GO:0070384) |
| 0.4 | 16.7 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.4 | 13.9 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.4 | 3.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.3 | 7.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 3.4 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.3 | 1.3 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.3 | 2.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.3 | 2.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.3 | 9.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 9.0 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.3 | 3.1 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.3 | 2.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.3 | 4.0 | GO:0035768 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.3 | 4.6 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.3 | 5.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 | 15.4 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.3 | 1.9 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.3 | 22.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 10.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 | 0.8 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.3 | 17.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 12.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 3.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.3 | 55.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.3 | 12.2 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.3 | 10.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.2 | 0.2 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.2 | 10.0 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 5.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 11.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 25.7 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.2 | 15.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 5.6 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 | 1.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 2.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.2 | 1.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.2 | 8.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 1.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 12.0 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.2 | 37.0 | GO:0009566 | fertilization(GO:0009566) |
| 0.2 | 11.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 0.8 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 11.8 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.2 | 8.9 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) |
| 0.2 | 0.9 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 31.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.2 | 0.5 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 0.7 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 23.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.2 | 8.5 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.2 | 9.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.2 | 3.7 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.2 | 4.3 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.2 | 9.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 2.8 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 | 2.6 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 8.2 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 2.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 2.7 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 7.1 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.7 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 16.0 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 8.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 2.9 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 1.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 5.6 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 5.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 2.3 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.1 | 9.3 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
| 0.1 | 6.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 3.8 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.1 | 0.3 | GO:0006186 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 1.3 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.3 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 7.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 25.0 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.1 | 1.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 3.2 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 0.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 2.9 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 3.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.8 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 23.1 | GO:0007411 | axon guidance(GO:0007411) |
| 0.1 | 1.5 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 2.0 | GO:0070875 | positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.1 | 3.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.1 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.1 | 0.5 | GO:0048549 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 1.7 | GO:0030534 | adult behavior(GO:0030534) |
| 0.1 | 0.2 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 2.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 5.0 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 0.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 4.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 2.5 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 2.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.0 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 1.7 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 1.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 3.9 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 1.4 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 0.3 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 1.7 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 1.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 6.5 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 4.4 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 5.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 4.1 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 16.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 1.1 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.2 | GO:0046782 | regulation of viral transcription(GO:0046782) positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 3.6 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.9 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 1.1 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 3.1 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 1.5 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 2.9 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 | 1.2 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 2.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.8 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 2.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 1.3 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 1.6 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.6 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 7.7 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.4 | 31.3 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 8.8 | 26.4 | GO:0001534 | radial spoke(GO:0001534) |
| 8.7 | 69.7 | GO:0002177 | manchette(GO:0002177) |
| 6.5 | 32.6 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 4.3 | 60.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 4.2 | 17.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 2.9 | 11.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 2.4 | 7.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 2.3 | 11.7 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.5 | 9.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.4 | 24.9 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 1.3 | 9.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 1.3 | 7.8 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.2 | 1.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 1.2 | 8.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 1.2 | 24.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 1.1 | 171.1 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 1.0 | 4.8 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.8 | 11.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.7 | 17.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.7 | 69.2 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.6 | 26.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.6 | 9.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.6 | 11.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.6 | 85.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.6 | 8.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.5 | 2.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.5 | 23.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.5 | 8.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.5 | 9.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.5 | 42.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.5 | 6.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.5 | 9.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 1.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.4 | 3.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.4 | 1.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 20.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 3.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.3 | 2.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 23.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.2 | 9.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 6.0 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 4.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 23.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 105.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.2 | 2.0 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 3.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 17.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 13.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 7.9 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 8.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.5 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 45.9 | GO:0005929 | cilium(GO:0005929) |
| 0.1 | 10.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 3.7 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 12.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 2.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 6.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.0 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 6.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 67.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 21.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 17.4 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 4.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 9.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.0 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 5.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 4.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 13.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.0 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 4.2 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 5.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 7.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 13.5 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 1.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 2.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 5.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.8 | GO:1902911 | protein kinase complex(GO:1902911) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 4.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 3.0 | 11.9 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 2.8 | 11.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 2.6 | 31.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.5 | 7.6 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 2.2 | 13.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 1.7 | 12.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 1.7 | 12.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 1.6 | 4.7 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 1.5 | 35.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 1.5 | 11.9 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 1.4 | 17.2 | GO:0071253 | connexin binding(GO:0071253) |
| 1.4 | 5.6 | GO:0004802 | transketolase activity(GO:0004802) |
| 1.4 | 4.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 1.3 | 1.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 1.2 | 15.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 1.2 | 3.5 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 1.0 | 10.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 1.0 | 19.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.0 | 26.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.8 | 21.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.8 | 22.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.8 | 3.9 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.8 | 3.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.8 | 11.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.7 | 7.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.7 | 8.7 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.7 | 23.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.7 | 17.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.7 | 36.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.7 | 52.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.7 | 2.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.7 | 20.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.6 | 3.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.6 | 7.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.6 | 9.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.5 | 12.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.5 | 9.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.5 | 13.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.5 | 5.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 1.5 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.5 | 4.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 13.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.4 | 3.9 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.4 | 8.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.4 | 8.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.4 | 151.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.4 | 8.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 12.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.4 | 5.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 4.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 2.8 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.3 | 1.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 2.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 9.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.3 | 2.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.3 | 2.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 19.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 9.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 2.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 21.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 2.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) phosphofructokinase activity(GO:0008443) |
| 0.3 | 6.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 9.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 3.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 7.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 1.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 1.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.2 | 12.8 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.2 | 1.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 6.5 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.2 | 10.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 11.0 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.2 | 0.7 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.2 | 6.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 26.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 4.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 13.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 1.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 2.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 3.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 15.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 22.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 1.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 27.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 7.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 3.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 7.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 14.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 1.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 7.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 8.7 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 12.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 23.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 64.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 33.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 2.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.3 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 2.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 1.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 3.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 3.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 1.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 18.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 8.0 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 1.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 6.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 2.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 1.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 4.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 1.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 4.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 3.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 2.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 11.5 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 5.4 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 1.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 16.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 15.3 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.8 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 28.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.6 | 44.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.4 | 20.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 11.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 11.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 16.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 8.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 10.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 11.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 5.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 8.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 3.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 7.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 5.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 2.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 7.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 32.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.9 | 26.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.9 | 40.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 11.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.6 | 18.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.5 | 21.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.5 | 2.8 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.3 | 14.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 18.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 5.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.2 | 6.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 4.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.2 | 13.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 9.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 28.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 25.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 6.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 8.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 6.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.6 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.1 | 4.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 4.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 28.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 3.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 12.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 3.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 5.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.7 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 1.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 7.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |