Project
Illumina Body Map 2
Navigation
Downloads
Results for RFX5
Z-value: 1.04
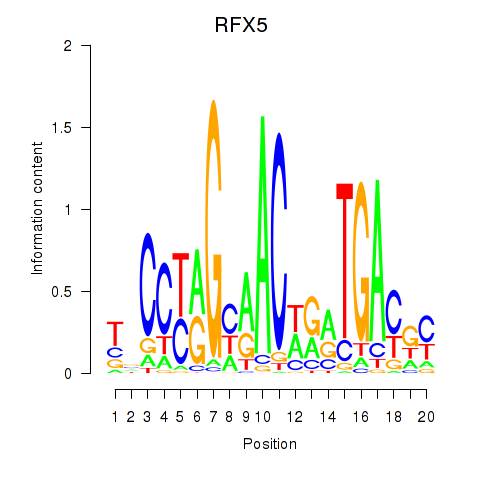
Transcription factors associated with RFX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX5
|
ENSG00000143390.13 | regulatory factor X5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX5 | hg19_v2_chr1_-_151319283_151319314 | 0.41 | 1.8e-02 | Click! |
Activity profile of RFX5 motif
Sorted Z-values of RFX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_142020496 | 4.98 |
ENST00000390381.3
|
TRBV5-1
|
T cell receptor beta variable 5-1 |
| chr6_-_32784687 | 4.59 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr22_+_44576770 | 4.56 |
ENST00000444313.3
ENST00000416291.1 |
PARVG
|
parvin, gamma |
| chr6_-_32731243 | 4.10 |
ENST00000427449.1
ENST00000411527.1 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr6_-_32498046 | 3.89 |
ENST00000374975.3
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5 |
| chr22_+_44577237 | 3.59 |
ENST00000415224.1
ENST00000417767.1 |
PARVG
|
parvin, gamma |
| chr6_-_32634425 | 3.58 |
ENST00000399082.3
ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr6_-_32731299 | 3.56 |
ENST00000435145.2
ENST00000437316.2 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr6_-_32908765 | 2.86 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr6_-_32908792 | 2.76 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr6_+_33043703 | 2.72 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr6_-_32557610 | 2.71 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr5_-_149792295 | 2.59 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr6_-_30658745 | 2.58 |
ENST00000376420.5
ENST00000376421.5 |
NRM
|
nurim (nuclear envelope membrane protein) |
| chr6_-_32920794 | 2.48 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr6_-_133079022 | 2.34 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr6_+_32812568 | 2.25 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr6_-_32821599 | 2.25 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr6_+_26383318 | 2.24 |
ENST00000469230.1
ENST00000490025.1 ENST00000356709.4 ENST00000352867.2 ENST00000493275.1 ENST00000472507.1 ENST00000482536.1 ENST00000432533.2 ENST00000482842.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr6_+_26383404 | 2.13 |
ENST00000416795.2
ENST00000494184.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr2_-_242088850 | 2.09 |
ENST00000358649.4
ENST00000452907.1 ENST00000403638.3 ENST00000539818.1 ENST00000415234.1 ENST00000234040.4 |
PASK
|
PAS domain containing serine/threonine kinase |
| chr6_-_32812420 | 1.92 |
ENST00000374881.2
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr6_+_32821924 | 1.80 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr13_-_47012325 | 1.65 |
ENST00000409879.2
|
KIAA0226L
|
KIAA0226-like |
| chr15_+_45003675 | 1.56 |
ENST00000558401.1
ENST00000559916.1 ENST00000544417.1 |
B2M
|
beta-2-microglobulin |
| chr6_+_26458171 | 1.47 |
ENST00000493173.1
ENST00000541522.1 ENST00000429381.1 ENST00000469185.1 |
BTN2A1
|
butyrophilin, subfamily 2, member A1 |
| chr4_-_143227088 | 1.45 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr19_+_19496624 | 1.42 |
ENST00000494516.2
ENST00000360315.3 ENST00000252577.5 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr6_+_26458152 | 1.42 |
ENST00000312541.5
|
BTN2A1
|
butyrophilin, subfamily 2, member A1 |
| chr4_-_143226979 | 1.41 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr1_+_181003067 | 1.28 |
ENST00000434571.2
ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1
|
major histocompatibility complex, class I-related |
| chr6_+_26365443 | 1.20 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr19_-_49118067 | 1.08 |
ENST00000593772.1
|
FAM83E
|
family with sequence similarity 83, member E |
| chr3_+_196466710 | 1.08 |
ENST00000327134.3
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr6_+_26365387 | 1.03 |
ENST00000532865.1
ENST00000530653.1 ENST00000527417.1 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr11_-_6640585 | 1.01 |
ENST00000533371.1
ENST00000528657.1 ENST00000436873.2 ENST00000299427.6 |
TPP1
|
tripeptidyl peptidase I |
| chr19_+_19496728 | 0.96 |
ENST00000537887.1
ENST00000417582.2 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr6_+_26440700 | 0.92 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr3_+_43328004 | 0.92 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr11_+_8008867 | 0.92 |
ENST00000309828.4
ENST00000449102.2 |
EIF3F
|
eukaryotic translation initiation factor 3, subunit F |
| chr1_+_249132462 | 0.90 |
ENST00000306562.3
|
ZNF672
|
zinc finger protein 672 |
| chr1_-_53608249 | 0.89 |
ENST00000371494.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr11_-_85430088 | 0.81 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr19_-_51014345 | 0.72 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr11_+_111385497 | 0.68 |
ENST00000375618.4
ENST00000529167.1 ENST00000332814.6 |
C11orf88
|
chromosome 11 open reading frame 88 |
| chr8_+_101170257 | 0.65 |
ENST00000251809.3
|
SPAG1
|
sperm associated antigen 1 |
| chr1_+_114473350 | 0.61 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr1_+_249132512 | 0.61 |
ENST00000505503.1
|
ZNF672
|
zinc finger protein 672 |
| chr16_+_67381263 | 0.53 |
ENST00000541146.1
ENST00000563189.1 ENST00000290940.7 |
LRRC36
|
leucine rich repeat containing 36 |
| chr17_+_27052892 | 0.50 |
ENST00000579671.1
ENST00000579060.1 |
NEK8
|
NIMA-related kinase 8 |
| chr19_-_6057282 | 0.50 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr9_+_43135095 | 0.48 |
ENST00000377437.2
|
AL513478.1
|
Uncharacterized protein |
| chr5_+_119799927 | 0.46 |
ENST00000407149.2
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr19_+_36103631 | 0.40 |
ENST00000203166.5
ENST00000379045.2 |
HAUS5
|
HAUS augmin-like complex, subunit 5 |
| chr1_-_208417620 | 0.33 |
ENST00000367033.3
|
PLXNA2
|
plexin A2 |
| chr7_+_766320 | 0.29 |
ENST00000297440.6
ENST00000313147.5 |
HEATR2
|
HEAT repeat containing 2 |
| chr16_+_67381289 | 0.28 |
ENST00000435835.3
|
LRRC36
|
leucine rich repeat containing 36 |
| chr9_-_67925210 | 0.27 |
ENST00000543078.1
|
BX649567.1
|
Uncharacterized protein |
| chr4_+_88529681 | 0.27 |
ENST00000399271.1
|
DSPP
|
dentin sialophosphoprotein |
| chr5_-_55412774 | 0.25 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr3_+_9851384 | 0.19 |
ENST00000419081.1
ENST00000438596.1 ENST00000417065.1 ENST00000439814.1 ENST00000418745.1 |
TTLL3
|
tubulin tyrosine ligase-like family, member 3 |
| chr12_-_123565834 | 0.13 |
ENST00000546049.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr10_+_82116529 | 0.12 |
ENST00000411538.1
ENST00000256039.2 |
DYDC2
|
DPY30 domain containing 2 |
| chr8_-_99306611 | 0.11 |
ENST00000341166.3
|
NIPAL2
|
NIPA-like domain containing 2 |
| chr10_+_14880287 | 0.07 |
ENST00000437161.2
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr17_+_30348024 | 0.06 |
ENST00000327564.7
ENST00000584368.1 ENST00000394713.3 ENST00000341671.7 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr7_-_15014398 | 0.06 |
ENST00000437998.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr10_+_14880157 | 0.01 |
ENST00000378372.3
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr22_-_30987837 | 0.01 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 10.8 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 1.5 | 4.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.8 | 1.6 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 0.6 | 2.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.5 | 2.6 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.3 | 2.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.2 | 4.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 | 1.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 2.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 17.8 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 0.3 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.1 | 2.2 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.8 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 5.8 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.6 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.9 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 2.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.3 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 7.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.5 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 2.4 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 1.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.7 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 35.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.7 | 6.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 2.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 1.6 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 0.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 1.3 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.1 | 0.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 2.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 2.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 15.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.5 | 2.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.5 | 2.9 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.4 | 2.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 11.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 8.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 6.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 1.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.8 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 1.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 8.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 15.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.1 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |