Project
Illumina Body Map 2
Navigation
Downloads
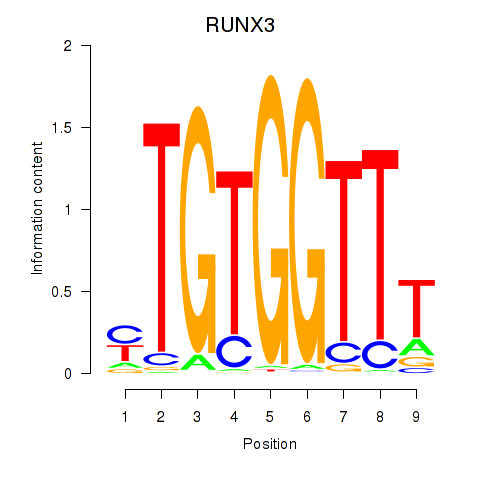
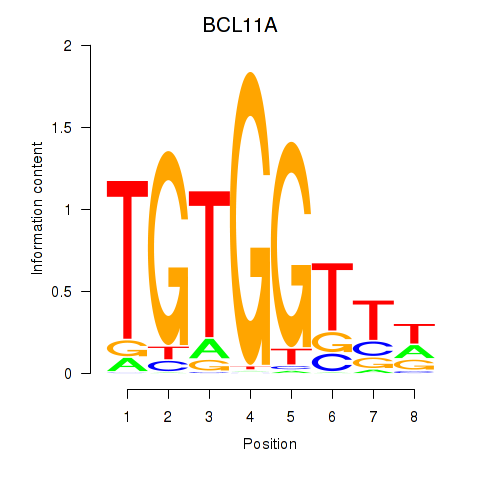
Results for RUNX3_BCL11A
Z-value: 3.13
Transcription factors associated with RUNX3_BCL11A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RUNX3
|
ENSG00000020633.14 | RUNX family transcription factor 3 |
|
BCL11A
|
ENSG00000119866.16 | BAF chromatin remodeling complex subunit BCL11A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RUNX3 | hg19_v2_chr1_-_25291475_25291511 | 0.91 | 5.9e-13 | Click! |
| BCL11A | hg19_v2_chr2_-_60780607_60780634 | 0.44 | 1.3e-02 | Click! |
Activity profile of RUNX3_BCL11A motif
Sorted Z-values of RUNX3_BCL11A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_40297079 | 23.84 |
ENST00000344138.4
ENST00000543252.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr3_-_150920979 | 23.08 |
ENST00000309180.5
ENST00000480322.1 |
GPR171
|
G protein-coupled receptor 171 |
| chr6_+_45296048 | 19.97 |
ENST00000465038.2
ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr1_-_114414316 | 19.04 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr5_+_156607829 | 18.76 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr22_+_40297105 | 18.08 |
ENST00000540310.1
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr2_-_158300556 | 17.25 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr1_-_25291475 | 16.38 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr4_+_71091786 | 15.38 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr2_+_7865923 | 14.98 |
ENST00000417930.1
|
AC092580.4
|
AC092580.4 |
| chr5_+_35856951 | 14.87 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr11_+_60739115 | 14.58 |
ENST00000344028.5
ENST00000346437.4 |
CD6
|
CD6 molecule |
| chr5_+_54398463 | 14.33 |
ENST00000274306.6
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr12_-_68553512 | 14.07 |
ENST00000229135.3
|
IFNG
|
interferon, gamma |
| chr11_+_60739140 | 13.87 |
ENST00000313421.7
|
CD6
|
CD6 molecule |
| chr11_+_60739249 | 13.83 |
ENST00000542157.1
ENST00000433107.2 ENST00000452451.2 ENST00000352009.5 |
CD6
|
CD6 molecule |
| chr17_+_7239821 | 12.97 |
ENST00000158762.3
ENST00000570457.2 |
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr14_-_25103472 | 12.74 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr19_-_42636617 | 12.66 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr5_+_133451254 | 12.48 |
ENST00000517851.1
ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr1_-_168513229 | 11.91 |
ENST00000367819.2
|
XCL2
|
chemokine (C motif) ligand 2 |
| chr14_-_25103388 | 11.44 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr17_+_7239904 | 11.07 |
ENST00000575425.1
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chrX_+_37639302 | 10.80 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr19_-_42636543 | 10.48 |
ENST00000528894.4
ENST00000560804.2 ENST00000560558.1 ENST00000560398.1 ENST00000526816.2 |
POU2F2
|
POU class 2 homeobox 2 |
| chrX_+_48542168 | 10.17 |
ENST00000376701.4
|
WAS
|
Wiskott-Aldrich syndrome |
| chr13_-_99910673 | 10.05 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chrX_-_49121165 | 9.95 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr3_+_114012819 | 9.79 |
ENST00000383671.3
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr1_+_168545711 | 9.48 |
ENST00000367818.3
|
XCL1
|
chemokine (C motif) ligand 1 |
| chr12_-_54694807 | 9.46 |
ENST00000435572.2
|
NFE2
|
nuclear factor, erythroid 2 |
| chr12_-_53594227 | 9.37 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chrX_-_70838306 | 9.10 |
ENST00000373691.4
ENST00000373693.3 |
CXCR3
|
chemokine (C-X-C motif) receptor 3 |
| chrX_+_37639264 | 9.01 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr3_+_98250743 | 8.92 |
ENST00000284311.3
|
GPR15
|
G protein-coupled receptor 15 |
| chr6_+_27925019 | 8.91 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr1_+_209929377 | 8.88 |
ENST00000400959.3
ENST00000367025.3 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr12_-_54694758 | 8.85 |
ENST00000553070.1
|
NFE2
|
nuclear factor, erythroid 2 |
| chr12_-_10542617 | 8.77 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr1_+_209929494 | 8.71 |
ENST00000367026.3
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr14_+_22337014 | 8.50 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr1_-_92951607 | 8.43 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr4_-_48116540 | 7.90 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr22_+_23222886 | 7.81 |
ENST00000390319.2
|
IGLV3-1
|
immunoglobulin lambda variable 3-1 |
| chr13_-_99910620 | 7.77 |
ENST00000416594.1
|
GPR18
|
G protein-coupled receptor 18 |
| chr1_+_209929446 | 7.69 |
ENST00000479796.1
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr2_+_90077680 | 7.32 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr4_-_109087906 | 7.30 |
ENST00000515500.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr17_-_34417479 | 7.27 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr19_-_8567505 | 7.22 |
ENST00000600262.1
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr9_+_75766763 | 7.19 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr6_+_45296391 | 7.15 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr20_+_44637526 | 7.14 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr19_-_50868882 | 6.98 |
ENST00000598915.1
|
NAPSA
|
napsin A aspartic peptidase |
| chr10_+_17272608 | 6.92 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr17_+_45810594 | 6.77 |
ENST00000177694.1
|
TBX21
|
T-box 21 |
| chr14_+_22236722 | 6.72 |
ENST00000390428.3
|
TRAV6
|
T cell receptor alpha variable 6 |
| chr7_+_18535321 | 6.67 |
ENST00000413380.1
ENST00000430454.1 |
HDAC9
|
histone deacetylase 9 |
| chr11_-_128457446 | 6.67 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr4_-_109087872 | 6.58 |
ENST00000510624.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr8_-_21771182 | 6.55 |
ENST00000523932.1
ENST00000544659.1 |
DOK2
|
docking protein 2, 56kDa |
| chr19_-_14785622 | 6.55 |
ENST00000443157.2
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr19_-_14785698 | 6.52 |
ENST00000344373.4
ENST00000595472.1 |
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr7_+_18535346 | 6.50 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr9_+_75766652 | 6.41 |
ENST00000257497.6
|
ANXA1
|
annexin A1 |
| chrX_-_19688475 | 6.38 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr17_-_29648761 | 6.36 |
ENST00000247270.3
ENST00000462804.2 |
EVI2A
|
ecotropic viral integration site 2A |
| chr1_+_53098862 | 6.31 |
ENST00000517870.1
|
FAM159A
|
family with sequence similarity 159, member A |
| chr1_+_221051699 | 6.29 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr17_-_61776522 | 6.25 |
ENST00000582055.1
|
LIMD2
|
LIM domain containing 2 |
| chr6_+_15401075 | 6.25 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr19_-_14785674 | 6.23 |
ENST00000253673.5
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr17_-_61777090 | 6.22 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr11_-_2323290 | 6.22 |
ENST00000381153.3
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr1_+_32716840 | 6.13 |
ENST00000336890.5
|
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr1_+_32716857 | 6.09 |
ENST00000482949.1
ENST00000495610.2 |
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr8_-_21771214 | 6.05 |
ENST00000276420.4
|
DOK2
|
docking protein 2, 56kDa |
| chr14_+_22465771 | 6.04 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr12_-_51717875 | 6.02 |
ENST00000604560.1
|
BIN2
|
bridging integrator 2 |
| chr3_-_72150076 | 5.98 |
ENST00000488545.1
ENST00000608654.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr19_-_8567478 | 5.88 |
ENST00000255612.3
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr8_+_22019168 | 5.88 |
ENST00000318561.3
ENST00000521315.1 ENST00000437090.2 ENST00000520605.1 ENST00000522109.1 ENST00000524255.1 ENST00000523296.1 ENST00000518615.1 |
SFTPC
|
surfactant protein C |
| chr12_-_51717948 | 5.88 |
ENST00000267012.4
|
BIN2
|
bridging integrator 2 |
| chr17_-_56494713 | 5.87 |
ENST00000407977.2
|
RNF43
|
ring finger protein 43 |
| chr8_-_21771173 | 5.77 |
ENST00000518197.1
|
DOK2
|
docking protein 2, 56kDa |
| chr12_-_51717922 | 5.76 |
ENST00000452142.2
|
BIN2
|
bridging integrator 2 |
| chr16_+_31366455 | 5.76 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr4_-_84035868 | 5.73 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr4_-_109087445 | 5.73 |
ENST00000512172.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr1_+_159770292 | 5.69 |
ENST00000536257.1
ENST00000321935.6 |
FCRL6
|
Fc receptor-like 6 |
| chr4_-_71532601 | 5.66 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr15_+_86098670 | 5.66 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr19_-_51875894 | 5.63 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr1_+_198608146 | 5.60 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr4_-_71532668 | 5.49 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr7_-_142176790 | 5.47 |
ENST00000390369.2
|
TRBV7-4
|
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr1_-_157522260 | 5.41 |
ENST00000368191.3
ENST00000361835.3 |
FCRL5
|
Fc receptor-like 5 |
| chr4_-_71532207 | 5.33 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr5_-_39270725 | 5.30 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr1_+_198608292 | 5.26 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr12_-_109025849 | 5.22 |
ENST00000228463.6
|
SELPLG
|
selectin P ligand |
| chr11_+_2323236 | 5.20 |
ENST00000182290.4
|
TSPAN32
|
tetraspanin 32 |
| chr1_-_157522180 | 5.20 |
ENST00000356953.4
ENST00000368188.2 ENST00000368190.3 ENST00000368189.3 |
FCRL5
|
Fc receptor-like 5 |
| chr4_-_84035905 | 5.17 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr15_+_58430368 | 5.15 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr4_-_71532339 | 5.15 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr19_-_36233332 | 5.12 |
ENST00000592537.1
ENST00000246532.1 ENST00000344990.3 ENST00000588992.1 |
IGFLR1
|
IGF-like family receptor 1 |
| chr1_+_172628154 | 5.12 |
ENST00000340030.3
ENST00000367721.2 |
FASLG
|
Fas ligand (TNF superfamily, member 6) |
| chr11_+_2323349 | 5.06 |
ENST00000381121.3
|
TSPAN32
|
tetraspanin 32 |
| chr3_+_111260980 | 5.00 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr17_-_47287928 | 4.92 |
ENST00000507680.1
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr3_+_113251143 | 4.76 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr3_+_111260954 | 4.74 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr3_+_111260856 | 4.74 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr17_-_73840614 | 4.68 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr17_-_73840415 | 4.64 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr2_+_103035102 | 4.62 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr11_+_35198243 | 4.61 |
ENST00000528455.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_-_207095212 | 4.60 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr17_-_46262541 | 4.60 |
ENST00000579336.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr1_+_17634689 | 4.60 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr14_+_22508822 | 4.59 |
ENST00000390448.3
|
TRAV20
|
T cell receptor alpha variable 20 |
| chr15_+_58430567 | 4.58 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr3_-_151047327 | 4.52 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr17_+_38673270 | 4.51 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr11_-_118122996 | 4.51 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr11_-_2323089 | 4.47 |
ENST00000456145.2
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr11_-_58345569 | 4.45 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr1_+_26872324 | 4.42 |
ENST00000531382.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr11_+_35198118 | 4.39 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr10_+_17270214 | 4.33 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr19_+_7413835 | 4.31 |
ENST00000576789.1
|
CTB-133G6.1
|
CTB-133G6.1 |
| chr12_-_53012343 | 4.28 |
ENST00000305748.3
|
KRT73
|
keratin 73 |
| chr6_+_31543334 | 4.24 |
ENST00000449264.2
|
TNF
|
tumor necrosis factor |
| chr6_+_144904334 | 4.21 |
ENST00000367526.4
|
UTRN
|
utrophin |
| chr5_-_94417562 | 4.20 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr5_-_138861926 | 4.16 |
ENST00000510817.1
|
TMEM173
|
transmembrane protein 173 |
| chr19_-_44285401 | 4.14 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr13_-_46756351 | 4.12 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr6_+_30029008 | 4.06 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr8_+_24241789 | 4.05 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr1_-_153517473 | 4.04 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr1_-_153348067 | 4.02 |
ENST00000368737.3
|
S100A12
|
S100 calcium binding protein A12 |
| chr16_+_85942594 | 4.01 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr3_-_112218378 | 4.01 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr1_+_158223923 | 3.96 |
ENST00000289429.5
|
CD1A
|
CD1a molecule |
| chr5_-_39203093 | 3.94 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr1_-_207095324 | 3.94 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr8_-_60031762 | 3.93 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr7_+_18330035 | 3.92 |
ENST00000413509.2
|
HDAC9
|
histone deacetylase 9 |
| chr3_-_112218205 | 3.84 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr14_+_22573582 | 3.83 |
ENST00000390453.1
|
TRAV24
|
T cell receptor alpha variable 24 |
| chr11_+_1892102 | 3.81 |
ENST00000417766.1
|
LSP1
|
lymphocyte-specific protein 1 |
| chr20_+_54987168 | 3.81 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr1_+_113161778 | 3.76 |
ENST00000263168.3
|
CAPZA1
|
capping protein (actin filament) muscle Z-line, alpha 1 |
| chr1_-_161039456 | 3.74 |
ENST00000368016.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr14_+_75988768 | 3.70 |
ENST00000286639.6
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr2_-_175462456 | 3.69 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr6_-_42418999 | 3.67 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr17_-_34524157 | 3.58 |
ENST00000378354.4
ENST00000394484.1 |
CCL3L3
|
chemokine (C-C motif) ligand 3-like 3 |
| chr17_-_34625719 | 3.53 |
ENST00000422211.2
ENST00000542124.1 |
CCL3L1
|
chemokine (C-C motif) ligand 3-like 1 |
| chr7_-_115670792 | 3.53 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr14_+_75988851 | 3.51 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr3_-_72149553 | 3.51 |
ENST00000468646.2
ENST00000464271.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr5_-_94417314 | 3.48 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr8_-_134072593 | 3.45 |
ENST00000427060.2
|
SLA
|
Src-like-adaptor |
| chr15_-_55563072 | 3.38 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr9_-_130541017 | 3.35 |
ENST00000314830.8
|
SH2D3C
|
SH2 domain containing 3C |
| chr2_+_30455016 | 3.33 |
ENST00000401506.1
ENST00000407930.2 |
LBH
|
limb bud and heart development |
| chr17_+_47287749 | 3.32 |
ENST00000419580.2
|
ABI3
|
ABI family, member 3 |
| chr1_-_145715565 | 3.28 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chr5_-_94417186 | 3.28 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr20_+_36932521 | 3.27 |
ENST00000262865.4
|
BPI
|
bactericidal/permeability-increasing protein |
| chr22_-_30642728 | 3.27 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr2_-_175462934 | 3.19 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr7_-_115670804 | 3.17 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr8_+_24241969 | 3.15 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr1_-_235098935 | 3.15 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr1_+_117544366 | 3.15 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr15_-_60695071 | 3.12 |
ENST00000557904.1
|
ANXA2
|
annexin A2 |
| chr8_-_101718991 | 3.11 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr17_-_1532106 | 3.11 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr5_+_148206156 | 3.08 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chr20_+_54987305 | 3.04 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr4_-_54930790 | 3.01 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr1_-_206945830 | 3.00 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr12_+_54410664 | 2.99 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr1_-_161039753 | 2.99 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chrX_+_155227246 | 2.93 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chr5_-_94417339 | 2.92 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr11_+_36397528 | 2.91 |
ENST00000311599.5
ENST00000378867.3 |
PRR5L
|
proline rich 5 like |
| chr8_-_101719159 | 2.85 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_-_103889731 | 2.81 |
ENST00000552578.1
ENST00000548048.1 ENST00000315192.8 ENST00000548883.1 ENST00000378113.2 |
C12orf42
|
chromosome 12 open reading frame 42 |
| chr15_+_49715293 | 2.78 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr13_-_28674693 | 2.77 |
ENST00000537084.1
ENST00000241453.7 ENST00000380982.4 |
FLT3
|
fms-related tyrosine kinase 3 |
| chr1_+_111772375 | 2.76 |
ENST00000467038.2
ENST00000497587.2 |
CHI3L2
|
chitinase 3-like 2 |
| chrX_+_47441712 | 2.71 |
ENST00000218388.4
ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr19_+_10947251 | 2.70 |
ENST00000592854.1
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr15_-_55562479 | 2.68 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chrX_+_155227371 | 2.66 |
ENST00000369423.2
ENST00000540897.1 |
IL9R
|
interleukin 9 receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of RUNX3_BCL11A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 24.0 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 4.8 | 14.5 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 4.8 | 19.0 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 4.5 | 13.6 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 4.5 | 17.8 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 4.0 | 19.8 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 3.2 | 9.5 | GO:2000562 | positive regulation of thymocyte migration(GO:2000412) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 3.0 | 24.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 2.9 | 8.8 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 2.8 | 8.4 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 2.8 | 19.6 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 2.7 | 13.3 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 2.5 | 10.2 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 2.4 | 4.9 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 2.3 | 9.3 | GO:0002432 | granuloma formation(GO:0002432) |
| 2.2 | 10.9 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 2.1 | 40.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 2.1 | 4.2 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 2.1 | 27.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.9 | 9.4 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 1.8 | 12.5 | GO:0048619 | embryonic genitalia morphogenesis(GO:0030538) embryonic hindgut morphogenesis(GO:0048619) |
| 1.6 | 9.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 1.6 | 11.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.4 | 4.2 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 1.3 | 6.7 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 1.3 | 5.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 1.2 | 14.9 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 1.2 | 7.3 | GO:2000503 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.2 | 4.7 | GO:2001301 | cellular response to interleukin-13(GO:0035963) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 1.2 | 4.7 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 1.1 | 7.9 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 1.1 | 9.0 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.1 | 18.8 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 1.0 | 3.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 1.0 | 14.3 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 1.0 | 7.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 1.0 | 28.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 1.0 | 13.1 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 1.0 | 7.0 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 1.0 | 21.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 1.0 | 4.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.9 | 5.7 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.9 | 5.5 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.9 | 4.6 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.9 | 6.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.9 | 2.6 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.9 | 5.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) endosomal lumen acidification(GO:0048388) |
| 0.8 | 24.1 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.8 | 11.3 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.8 | 3.1 | GO:0052214 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.8 | 16.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.8 | 0.8 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.7 | 2.9 | GO:0002884 | negative regulation of type IV hypersensitivity(GO:0001808) negative regulation of hypersensitivity(GO:0002884) |
| 0.7 | 3.5 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.7 | 5.6 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.7 | 2.0 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.6 | 1.9 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.6 | 1.9 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.6 | 2.5 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.6 | 2.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.6 | 1.7 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.5 | 10.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.5 | 18.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.5 | 3.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.5 | 5.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.5 | 6.0 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.5 | 4.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.5 | 3.4 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.5 | 6.8 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.5 | 24.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.5 | 11.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.4 | 6.7 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.4 | 8.9 | GO:0072678 | T cell migration(GO:0072678) |
| 0.4 | 4.4 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.4 | 51.6 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.4 | 7.8 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 9.8 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.4 | 3.3 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.4 | 1.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.4 | 1.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.4 | 2.4 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.4 | 4.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.4 | 2.8 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.4 | 20.9 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.4 | 1.2 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.4 | 6.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 5.5 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.4 | 2.7 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.4 | 1.2 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.4 | 1.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.3 | 1.4 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.3 | 1.4 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.3 | 1.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.3 | 4.8 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 2.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 1.1 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.3 | 0.8 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 0.8 | GO:1901656 | cellular response to mycotoxin(GO:0036146) glycoside transport(GO:1901656) cellular response to bile acid(GO:1903413) |
| 0.2 | 1.0 | GO:0060558 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) regulation of calcidiol 1-monooxygenase activity(GO:0060558) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.2 | 0.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 4.2 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.2 | 1.2 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.2 | 4.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 0.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 4.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 1.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 10.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.2 | 0.4 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.2 | 1.3 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.2 | 5.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 1.5 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 3.3 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.2 | 3.0 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.2 | 0.5 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.2 | 1.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.5 | GO:0043132 | NAD transport(GO:0043132) |
| 0.1 | 1.7 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 4.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 24.6 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 1.3 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.9 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 2.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.3 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.1 | 8.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 2.1 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.1 | 2.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.3 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.1 | 3.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 3.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 1.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 2.3 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 3.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.5 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.1 | 0.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 1.6 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.4 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 7.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 5.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.7 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.3 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 4.0 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 1.5 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 4.7 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 1.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 2.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.9 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 6.4 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.1 | 4.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.5 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.3 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 5.8 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 1.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.3 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.5 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.1 | 0.7 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 1.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.2 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.1 | 1.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.5 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 1.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 1.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.8 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 4.9 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.6 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 8.9 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 4.7 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.8 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 2.1 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.6 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 7.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.8 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.7 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 1.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 16.4 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 18.3 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.6 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 3.0 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 1.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 1.7 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 1.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.3 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 1.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 3.3 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.4 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.4 | GO:0051131 | trophectodermal cell differentiation(GO:0001829) chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 4.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.9 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.4 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.4 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 1.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.4 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 1.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 1.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.9 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 2.1 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.2 | GO:0002076 | osteoblast development(GO:0002076) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.4 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 1.5 | 13.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.4 | 19.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 1.2 | 93.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.1 | 9.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.1 | 12.7 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 1.0 | 19.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.9 | 19.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.9 | 26.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.8 | 3.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.7 | 3.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.5 | 3.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.5 | 2.1 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.5 | 6.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.4 | 89.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.3 | 6.5 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 0.8 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 5.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 11.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 23.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 12.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 4.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 30.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 5.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 5.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 4.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 4.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 62.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 4.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 4.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 10.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 20.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 14.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 10.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 5.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 18.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 10.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 1.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 43.2 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 1.0 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 8.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 3.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.5 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 1.2 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 5.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.3 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 1.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 1.7 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 19.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 9.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 22.1 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 2.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 3.9 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 1.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.7 | GO:0097223 | acrosomal vesicle(GO:0001669) sperm part(GO:0097223) |
| 0.0 | 1.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.5 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.0 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 42.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 3.7 | 14.9 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 1.9 | 9.7 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 1.6 | 14.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.6 | 11.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.6 | 4.8 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 1.5 | 16.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.2 | 7.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 1.2 | 4.7 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 1.1 | 6.7 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.0 | 19.6 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 1.0 | 3.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 1.0 | 28.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.9 | 5.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.9 | 4.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.9 | 2.6 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.8 | 19.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.8 | 9.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.7 | 75.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.7 | 4.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.7 | 2.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.7 | 4.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.7 | 2.0 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.6 | 1.9 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.6 | 21.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.6 | 24.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.6 | 17.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.6 | 9.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.5 | 5.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.4 | 7.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.4 | 1.6 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.4 | 4.0 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.4 | 1.2 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.4 | 3.5 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.4 | 3.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.4 | 27.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.4 | 1.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.4 | 5.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 8.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 3.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.3 | 10.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 1.0 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.3 | 11.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 1.6 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 6.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 3.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 14.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 2.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.3 | 3.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.2 | 1.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.2 | 5.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.3 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.2 | 8.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 6.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.5 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.2 | 9.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 4.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 6.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 10.8 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.2 | 1.2 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.2 | 12.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 4.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 0.7 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 1.7 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.2 | 2.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 3.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 2.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.2 | 8.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 1.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 1.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 6.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 2.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 2.3 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.4 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 1.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 4.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 15.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 5.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 44.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 3.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.3 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 12.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.4 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 3.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 13.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 4.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 4.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 4.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 1.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 9.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 37.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 15.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 8.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 4.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 1.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 1.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 1.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 13.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.5 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 1.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 14.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 3.8 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 1.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 4.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.9 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 9.3 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 6.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 2.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 5.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.1 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 2.4 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.0 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 1.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 41.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.9 | 14.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.7 | 29.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.6 | 7.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.5 | 23.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.5 | 64.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.5 | 9.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.5 | 5.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.5 | 15.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.4 | 29.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.4 | 39.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.3 | 27.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 10.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.3 | 29.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 9.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 32.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 19.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 22.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 8.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 5.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 7.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 9.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 5.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 1.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 29.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 6.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 5.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 5.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 4.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 7.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 24.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 5.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 3.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 6.8 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 92.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 1.4 | 10.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.6 | 9.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.6 | 46.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.5 | 18.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.5 | 14.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.5 | 27.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 14.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.4 | 9.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 5.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 10.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 23.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.3 | 35.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 11.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 11.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 7.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 13.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 7.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 5.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 21.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 5.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 5.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 32.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 3.9 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.2 | 6.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 21.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 7.4 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.1 | 7.9 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 6.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 1.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 4.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 18.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 7.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 18.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 4.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 1.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 3.0 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 2.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.9 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 2.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 2.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |