Project
Illumina Body Map 2
Navigation
Downloads
Results for SHOX
Z-value: 1.46
Transcription factors associated with SHOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SHOX
|
ENSG00000185960.8 | short stature homeobox |
|
SHOX
|
ENSGR0000185960.8 | short stature homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SHOX | hg19_v2_chrX_+_591524_591542 | 0.32 | 7.7e-02 | Click! |
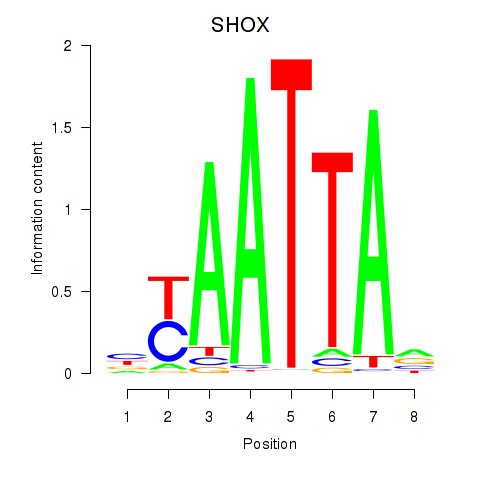
Activity profile of SHOX motif
Sorted Z-values of SHOX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_36245561 | 6.53 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr13_-_46716969 | 6.44 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_-_92952433 | 6.40 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr3_-_27764190 | 6.05 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr3_-_72149553 | 5.43 |
ENST00000468646.2
ENST00000464271.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr18_-_44181442 | 4.91 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr3_-_27763803 | 4.72 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr4_-_25865159 | 4.70 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr15_+_58430567 | 4.30 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr2_+_90248739 | 4.11 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr15_+_58430368 | 4.09 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr7_+_50348268 | 4.01 |
ENST00000438033.1
ENST00000439701.1 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr4_-_57547870 | 3.96 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr5_-_24645078 | 3.83 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr4_+_40201954 | 3.77 |
ENST00000511121.1
|
RHOH
|
ras homolog family member H |
| chr1_-_183538319 | 3.60 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr12_-_10282742 | 3.59 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr6_-_9933500 | 3.55 |
ENST00000492169.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr1_+_40713573 | 3.55 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr14_+_22309368 | 3.50 |
ENST00000390433.1
|
TRAV12-1
|
T cell receptor alpha variable 12-1 |
| chr4_-_57547454 | 3.49 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr14_+_22771851 | 3.42 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chr3_-_191000172 | 3.42 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chrX_+_134654540 | 3.36 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr16_+_12059050 | 3.35 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr6_+_135502501 | 3.30 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr12_-_10282836 | 3.30 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr12_-_10282681 | 3.26 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr6_-_32157947 | 3.18 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chrX_+_107288280 | 3.12 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_-_72149448 | 3.08 |
ENST00000498432.1
|
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr17_-_78450398 | 3.06 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr11_-_117748138 | 3.02 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr13_-_99910620 | 3.01 |
ENST00000416594.1
|
GPR18
|
G protein-coupled receptor 18 |
| chr17_-_38956205 | 3.00 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr11_-_117747607 | 2.94 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr12_-_91398796 | 2.78 |
ENST00000261172.3
ENST00000551767.1 |
EPYC
|
epiphycan |
| chr7_+_142378566 | 2.71 |
ENST00000390398.3
|
TRBV25-1
|
T cell receptor beta variable 25-1 |
| chr15_+_75080883 | 2.70 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr2_+_169926047 | 2.69 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr6_-_49681235 | 2.68 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr17_-_72527605 | 2.67 |
ENST00000392621.1
ENST00000314401.3 |
CD300LB
|
CD300 molecule-like family member b |
| chr7_+_37723336 | 2.65 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr11_-_117747434 | 2.62 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr3_-_167191814 | 2.58 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr14_+_22670455 | 2.57 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr2_-_145278475 | 2.51 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr13_-_95131923 | 2.50 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chrX_+_135730373 | 2.48 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr6_-_116866773 | 2.47 |
ENST00000368602.3
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr11_+_128563652 | 2.44 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr18_+_57567180 | 2.43 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr1_+_198608146 | 2.43 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr3_+_115342349 | 2.38 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr12_+_8666126 | 2.38 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr14_+_22739823 | 2.37 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr1_+_50569575 | 2.36 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr4_-_143227088 | 2.36 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr12_-_10605929 | 2.33 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr4_-_143226979 | 2.28 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr1_+_198607801 | 2.27 |
ENST00000367379.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr7_-_92777606 | 2.27 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr1_+_222910625 | 2.27 |
ENST00000360827.2
|
FAM177B
|
family with sequence similarity 177, member B |
| chr2_+_152214098 | 2.26 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr3_+_141106643 | 2.25 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr7_+_115862858 | 2.25 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr5_+_118668846 | 2.24 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr17_-_38911580 | 2.24 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr15_+_86087267 | 2.24 |
ENST00000558166.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr11_-_85430356 | 2.22 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr10_-_56561022 | 2.22 |
ENST00000373965.2
ENST00000414778.1 ENST00000395438.1 ENST00000409834.1 ENST00000395445.1 ENST00000395446.1 ENST00000395442.1 ENST00000395440.1 ENST00000395432.2 ENST00000361849.3 ENST00000395433.1 ENST00000320301.6 ENST00000395430.1 ENST00000437009.1 |
PCDH15
|
protocadherin-related 15 |
| chrX_-_19817869 | 2.22 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr2_-_89327228 | 2.21 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr1_+_198608292 | 2.21 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr11_-_15643937 | 2.18 |
ENST00000533082.1
|
RP11-531H8.2
|
RP11-531H8.2 |
| chr13_+_78315295 | 2.16 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_+_107682629 | 2.12 |
ENST00000370074.4
ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1
|
netrin G1 |
| chr19_-_7698599 | 2.11 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr1_+_206557366 | 2.10 |
ENST00000414007.1
ENST00000419187.2 |
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr19_-_47137942 | 2.10 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr5_-_121659052 | 2.09 |
ENST00000512105.1
|
CTD-2544H17.1
|
CTD-2544H17.1 |
| chr21_-_42219065 | 2.08 |
ENST00000400454.1
|
DSCAM
|
Down syndrome cell adhesion molecule |
| chr1_-_150738261 | 2.07 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr12_+_10163231 | 2.06 |
ENST00000396502.1
ENST00000338896.5 |
CLEC12B
|
C-type lectin domain family 12, member B |
| chr5_-_141030943 | 2.05 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr12_+_10124110 | 2.04 |
ENST00000350667.4
|
CLEC12A
|
C-type lectin domain family 12, member A |
| chr9_-_215744 | 2.04 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr13_+_78109884 | 2.04 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr6_+_31583761 | 2.04 |
ENST00000376049.4
|
AIF1
|
allograft inflammatory factor 1 |
| chr17_-_9929581 | 2.03 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr17_+_61151306 | 2.03 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr11_+_4470525 | 2.02 |
ENST00000325719.4
|
OR52K2
|
olfactory receptor, family 52, subfamily K, member 2 |
| chr2_-_203735484 | 2.02 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr12_-_14133053 | 2.02 |
ENST00000609686.1
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| chr1_-_157746909 | 2.01 |
ENST00000392274.3
ENST00000361516.3 ENST00000368181.4 |
FCRL2
|
Fc receptor-like 2 |
| chr4_-_116034979 | 2.01 |
ENST00000264363.2
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chrX_-_13835147 | 2.00 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chrX_+_107288197 | 2.00 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr12_+_41136144 | 2.00 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr9_+_125132803 | 1.99 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr6_-_161695042 | 1.98 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr10_+_111765562 | 1.98 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr2_+_68961934 | 1.97 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr9_+_44867571 | 1.96 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr13_+_78315348 | 1.95 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr9_+_125133315 | 1.95 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr14_+_65016620 | 1.95 |
ENST00000298705.1
|
PPP1R36
|
protein phosphatase 1, regulatory subunit 36 |
| chr3_+_121774202 | 1.93 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr2_+_204571375 | 1.93 |
ENST00000374478.4
|
CD28
|
CD28 molecule |
| chr3_+_111393501 | 1.92 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr12_-_89746264 | 1.92 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr2_-_145277569 | 1.91 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr18_-_30353025 | 1.91 |
ENST00000359358.4
|
KLHL14
|
kelch-like family member 14 |
| chr8_+_24298597 | 1.91 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr4_-_109541610 | 1.90 |
ENST00000510212.1
|
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr9_-_123639445 | 1.89 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr2_+_68961905 | 1.88 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr12_-_89746173 | 1.86 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr12_-_31478428 | 1.86 |
ENST00000543615.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr12_+_4385230 | 1.86 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr13_+_78109804 | 1.86 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr1_-_242612779 | 1.85 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr6_-_161695074 | 1.85 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr2_+_89184868 | 1.83 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr9_-_16728161 | 1.82 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chrX_+_107288239 | 1.80 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr13_-_41593425 | 1.80 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr4_+_69313145 | 1.78 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr18_+_21572737 | 1.78 |
ENST00000304621.6
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr8_-_42234745 | 1.78 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr1_+_66458072 | 1.77 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr2_+_162272605 | 1.77 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr8_+_24298531 | 1.77 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr1_+_107683436 | 1.77 |
ENST00000370068.1
|
NTNG1
|
netrin G1 |
| chr10_+_90660832 | 1.76 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chrX_-_153602991 | 1.76 |
ENST00000369850.3
ENST00000422373.1 |
FLNA
|
filamin A, alpha |
| chr5_+_96840389 | 1.75 |
ENST00000504012.1
|
RP11-1E3.1
|
RP11-1E3.1 |
| chr15_-_98836406 | 1.74 |
ENST00000560360.1
|
CTD-2544M6.1
|
CTD-2544M6.1 |
| chr6_+_105404899 | 1.74 |
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans) |
| chr9_+_82186872 | 1.73 |
ENST00000376544.3
ENST00000376520.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr6_-_39693111 | 1.73 |
ENST00000373215.3
ENST00000538893.1 ENST00000287152.7 ENST00000373216.3 |
KIF6
|
kinesin family member 6 |
| chr20_+_42523336 | 1.73 |
ENST00000428529.1
|
RP5-1030M6.3
|
RP5-1030M6.3 |
| chr1_+_244515930 | 1.72 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr7_+_129984630 | 1.72 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr1_+_117544366 | 1.71 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr4_-_41884620 | 1.71 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr8_+_50824233 | 1.71 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr2_-_224467002 | 1.71 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr20_+_3776371 | 1.71 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr2_+_90211643 | 1.68 |
ENST00000390277.2
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr1_+_168250194 | 1.67 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr2_+_90273679 | 1.66 |
ENST00000423080.2
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr3_-_164796269 | 1.64 |
ENST00000264382.3
|
SI
|
sucrase-isomaltase (alpha-glucosidase) |
| chr14_+_72052983 | 1.63 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr19_-_51522955 | 1.63 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr3_-_108248169 | 1.62 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr13_-_36788718 | 1.62 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr2_+_68962014 | 1.61 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr7_-_78400364 | 1.60 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_-_116833500 | 1.60 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chrX_-_153599578 | 1.57 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr4_+_78432907 | 1.57 |
ENST00000286758.4
|
CXCL13
|
chemokine (C-X-C motif) ligand 13 |
| chr2_+_78143006 | 1.56 |
ENST00000443419.1
|
AC073628.1
|
AC073628.1 |
| chr9_+_82188077 | 1.56 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr18_-_31803435 | 1.55 |
ENST00000589544.1
ENST00000269185.4 ENST00000261592.5 |
NOL4
|
nucleolar protein 4 |
| chr18_+_22040620 | 1.54 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr4_-_123542224 | 1.51 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chrX_+_123097014 | 1.51 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr7_-_92747269 | 1.51 |
ENST00000446617.1
ENST00000379958.2 |
SAMD9
|
sterile alpha motif domain containing 9 |
| chr7_-_28220354 | 1.51 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr20_+_15177480 | 1.51 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr14_-_36988882 | 1.50 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr6_+_130339710 | 1.50 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr4_-_66536057 | 1.48 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr14_+_59100774 | 1.46 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr13_+_78315528 | 1.46 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr6_+_155538093 | 1.44 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr13_+_78315466 | 1.43 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr5_+_179159813 | 1.42 |
ENST00000292599.3
|
MAML1
|
mastermind-like 1 (Drosophila) |
| chr2_-_61697862 | 1.42 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr6_-_32908792 | 1.42 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr12_-_94673956 | 1.41 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr20_+_18488137 | 1.40 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr17_+_57233087 | 1.40 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr2_+_17997763 | 1.40 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr1_+_225600404 | 1.40 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr3_+_111718036 | 1.39 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr20_+_2276639 | 1.39 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr2_-_80531720 | 1.38 |
ENST00000416268.1
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr15_-_55562582 | 1.38 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_+_103035102 | 1.38 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr14_-_98444438 | 1.37 |
ENST00000512901.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr2_-_80531399 | 1.36 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr18_+_50278430 | 1.35 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chrX_+_22050546 | 1.35 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr8_+_38831683 | 1.35 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr3_-_151034734 | 1.34 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr16_+_12059091 | 1.33 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr15_-_55562479 | 1.32 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_-_185597619 | 1.31 |
ENST00000608417.1
ENST00000436955.1 |
GS1-204I12.1
|
GS1-204I12.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SHOX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 2.1 | 6.4 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 1.4 | 8.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 1.4 | 6.9 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 1.1 | 3.3 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.1 | 3.3 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.8 | 3.8 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.8 | 3.0 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.7 | 5.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.6 | 1.9 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.6 | 2.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.5 | 1.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.5 | 2.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.5 | 4.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.5 | 10.5 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.5 | 1.4 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.4 | 1.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.4 | 1.7 | GO:0021536 | diencephalon development(GO:0021536) pituitary gland development(GO:0021983) |
| 0.4 | 1.6 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.4 | 1.6 | GO:2001027 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.4 | 1.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.4 | 2.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.4 | 2.5 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.4 | 1.4 | GO:0060928 | atrioventricular node development(GO:0003162) cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.3 | 1.4 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.3 | 1.3 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.3 | 2.0 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.3 | 1.0 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.3 | 1.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.3 | 2.7 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.3 | 1.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.3 | 1.8 | GO:0021764 | amygdala development(GO:0021764) |
| 0.3 | 1.8 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.3 | 2.0 | GO:0071672 | regulation of muscle hyperplasia(GO:0014738) negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.3 | 7.7 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.3 | 2.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.3 | 2.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 0.7 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 3.5 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 6.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 2.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.0 | GO:0042414 | epinephrine metabolic process(GO:0042414) cellular response to lead ion(GO:0071284) |
| 0.2 | 6.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 1.6 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 5.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.3 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.2 | 1.8 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.2 | 0.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.5 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 0.8 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.2 | 1.4 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.2 | 2.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 1.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 2.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.2 | 0.7 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.2 | 2.0 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 2.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 4.6 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.2 | 1.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 0.8 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 1.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 2.4 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.2 | 2.7 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 3.8 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.2 | 3.8 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 0.6 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 1.2 | GO:1901295 | positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 2.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 1.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 1.9 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.7 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.4 | GO:1905132 | metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 1.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.9 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 1.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.5 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.7 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.7 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.9 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.1 | 0.6 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.1 | 0.9 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 2.5 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 2.0 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 3.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 2.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 2.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 2.0 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.8 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 2.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 3.6 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 1.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.2 | GO:0060022 | hard palate development(GO:0060022) |
| 0.1 | 5.9 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 1.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 5.6 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 2.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 1.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 1.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 8.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 1.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.8 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 0.6 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 1.4 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 1.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 2.2 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.1 | 0.6 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.4 | GO:0034959 | neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 1.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.9 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 1.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.8 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.1 | 2.0 | GO:0048584 | positive regulation of response to stimulus(GO:0048584) |
| 0.1 | 2.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 10.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 4.6 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 1.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 3.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 3.5 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 2.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.5 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 2.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.3 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 3.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 1.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.6 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.5 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 6.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.6 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 2.4 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
| 0.0 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 1.1 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 1.9 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.9 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 1.2 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 2.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 3.8 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 1.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 2.2 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 1.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 3.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.4 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 1.0 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 3.5 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 3.7 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 2.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 1.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 1.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.4 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.8 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 1.7 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 1.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.3 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 1.4 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.6 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.6 | GO:0031214 | biomineral tissue development(GO:0031214) |
| 0.0 | 1.7 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.3 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.8 | GO:0048477 | oogenesis(GO:0048477) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 3.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.3 | 2.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.3 | 2.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 0.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 8.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 4.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 1.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 4.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 2.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 3.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 0.6 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.2 | 2.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.7 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.2 | 2.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 0.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 1.8 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 2.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.5 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 7.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 3.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 3.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.7 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 5.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 5.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.8 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 8.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 6.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 3.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 7.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 7.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 1.0 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 4.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 8.8 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 11.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.3 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 3.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.2 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 2.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 4.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 2.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 2.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.3 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 2.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0031904 | endosome lumen(GO:0031904) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 1.0 | 5.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.8 | 2.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.8 | 4.6 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.5 | 3.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.5 | 2.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 1.8 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 2.5 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.4 | 1.6 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.4 | 1.6 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.4 | 2.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.4 | 2.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 1.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 1.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.3 | 10.5 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.3 | 1.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 1.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 1.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.2 | 4.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 2.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 3.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 2.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 2.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 3.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 2.7 | GO:0047035 | alcohol dehydrogenase (NAD) activity(GO:0004022) testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 1.7 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 1.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 0.5 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.2 | 6.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 2.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 0.6 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 2.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 1.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.8 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.8 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 1.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 1.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 3.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 3.8 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 3.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 2.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.1 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.1 | 7.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 4.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 8.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 1.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 2.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.7 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 5.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.6 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 3.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.5 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 5.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 1.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 14.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.6 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 1.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.2 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 1.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 2.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 2.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 6.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 1.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.7 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 6.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 4.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 2.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 2.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 2.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 4.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.8 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 1.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 4.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.7 | GO:0001948 | glycoprotein binding(GO:0001948) dystroglycan binding(GO:0002162) |
| 0.0 | 1.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 3.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.3 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 1.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 3.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0004871 | signal transducer activity(GO:0004871) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 4.4 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 5.7 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 1.7 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 13.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 8.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 4.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 5.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 6.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 19.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 2.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 2.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 4.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 2.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 3.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 4.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 9.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 9.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 4.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 2.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 4.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 3.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 3.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 5.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 3.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 6.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 19.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 2.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 4.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 2.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 3.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 3.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |