Project
Illumina Body Map 2
Navigation
Downloads
Results for SMAD2
Z-value: 1.23
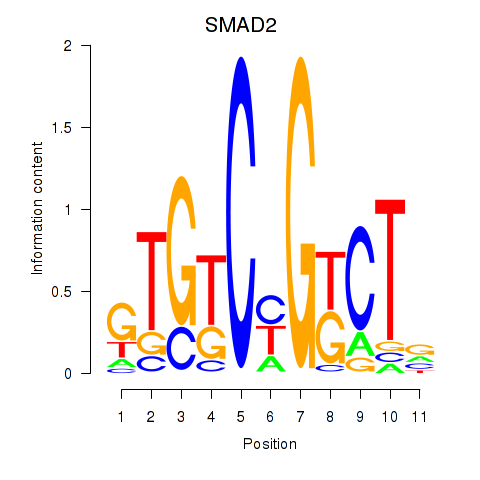
Transcription factors associated with SMAD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD2
|
ENSG00000175387.11 | SMAD family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD2 | hg19_v2_chr18_-_45457478_45457517 | 0.21 | 2.4e-01 | Click! |
Activity profile of SMAD2 motif
Sorted Z-values of SMAD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_159141397 | 3.84 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr7_-_100493482 | 2.91 |
ENST00000411582.1
ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr17_-_15165854 | 2.88 |
ENST00000395936.1
ENST00000395938.2 |
PMP22
|
peripheral myelin protein 22 |
| chr1_-_44482979 | 2.76 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_-_100493744 | 2.69 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr17_-_74533963 | 2.67 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr20_+_43343886 | 2.65 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr17_-_15165825 | 2.55 |
ENST00000426385.3
|
PMP22
|
peripheral myelin protein 22 |
| chr20_+_43343476 | 2.52 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr1_+_160085567 | 2.52 |
ENST00000392233.3
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr8_+_106331769 | 2.51 |
ENST00000520492.1
|
ZFPM2
|
zinc finger protein, FOG family member 2 |
| chrX_+_38420783 | 2.48 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr12_+_53443963 | 2.44 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chrX_+_38420623 | 2.42 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr1_+_160085501 | 2.40 |
ENST00000361216.3
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr11_-_107590383 | 2.39 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr1_-_201915590 | 2.37 |
ENST00000367288.4
|
LMOD1
|
leiomodin 1 (smooth muscle) |
| chr8_+_94241867 | 2.22 |
ENST00000598428.1
|
AC016885.1
|
Uncharacterized protein |
| chr19_-_46105411 | 2.20 |
ENST00000323040.4
ENST00000544371.1 |
GPR4
OPA3
|
G protein-coupled receptor 4 optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr21_+_47401650 | 2.14 |
ENST00000361866.3
|
COL6A1
|
collagen, type VI, alpha 1 |
| chr16_+_28889801 | 2.09 |
ENST00000395503.4
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr12_+_129178451 | 2.07 |
ENST00000537538.1
|
TMEM132C
|
transmembrane protein 132C |
| chr22_+_22723969 | 1.99 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr22_+_23010756 | 1.99 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chr20_+_43343517 | 1.98 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr2_-_72375167 | 1.89 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr11_-_107729887 | 1.84 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr11_-_119211525 | 1.84 |
ENST00000528368.1
|
C1QTNF5
|
C1q and tumor necrosis factor related protein 5 |
| chr16_+_28889703 | 1.81 |
ENST00000357084.3
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr14_-_106494587 | 1.80 |
ENST00000390597.2
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr1_+_160097462 | 1.79 |
ENST00000447527.1
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr1_-_44497118 | 1.79 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr16_-_70719925 | 1.78 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr7_-_102158157 | 1.77 |
ENST00000541662.1
ENST00000306682.6 ENST00000465829.1 |
RASA4B
|
RAS p21 protein activator 4B |
| chr9_+_71944241 | 1.76 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr7_-_102257139 | 1.72 |
ENST00000521076.1
ENST00000462172.1 ENST00000522801.1 ENST00000449970.2 ENST00000262940.7 |
RASA4
|
RAS p21 protein activator 4 |
| chr8_+_82192501 | 1.63 |
ENST00000297258.6
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated) |
| chr11_-_107729287 | 1.61 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr7_+_130020180 | 1.57 |
ENST00000481342.1
ENST00000011292.3 ENST00000604896.1 |
CPA1
|
carboxypeptidase A1 (pancreatic) |
| chr19_-_44285401 | 1.48 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr1_+_100111479 | 1.45 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chr3_+_123813509 | 1.41 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr9_-_136006496 | 1.38 |
ENST00000372062.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr11_-_107729504 | 1.38 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr6_+_32132360 | 1.37 |
ENST00000333845.6
ENST00000395512.1 ENST00000432129.1 |
EGFL8
|
EGF-like-domain, multiple 8 |
| chr20_+_30598231 | 1.31 |
ENST00000300415.8
ENST00000262659.8 |
CCM2L
|
cerebral cavernous malformation 2-like |
| chr14_+_31343747 | 1.29 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr1_+_100111580 | 1.26 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr12_+_53443680 | 1.25 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_-_10251539 | 1.25 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr10_+_72972281 | 1.24 |
ENST00000335350.6
|
UNC5B
|
unc-5 homolog B (C. elegans) |
| chr22_+_22676808 | 1.24 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr12_-_10251603 | 1.24 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr3_-_79068138 | 1.24 |
ENST00000495273.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr7_-_96654133 | 1.24 |
ENST00000486603.2
ENST00000222598.4 |
DLX5
|
distal-less homeobox 5 |
| chr12_-_22094336 | 1.23 |
ENST00000326684.4
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr14_-_106478603 | 1.20 |
ENST00000390596.2
|
IGHV4-4
|
immunoglobulin heavy variable 4-4 |
| chr4_-_5890145 | 1.19 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr22_+_23213658 | 1.16 |
ENST00000390318.2
|
IGLV4-3
|
immunoglobulin lambda variable 4-3 |
| chr5_-_172755056 | 1.13 |
ENST00000520648.1
|
STC2
|
stanniocalcin 2 |
| chr19_+_16999966 | 1.12 |
ENST00000599210.1
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr19_+_35521699 | 1.12 |
ENST00000415950.3
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr7_+_130020932 | 1.07 |
ENST00000484324.1
|
CPA1
|
carboxypeptidase A1 (pancreatic) |
| chr12_-_10251576 | 1.06 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr7_-_559853 | 1.06 |
ENST00000405692.2
|
PDGFA
|
platelet-derived growth factor alpha polypeptide |
| chr22_+_22735135 | 1.04 |
ENST00000390297.2
|
IGLV1-44
|
immunoglobulin lambda variable 1-44 |
| chr2_-_220174166 | 1.03 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr21_-_36259445 | 1.01 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr16_+_31483374 | 0.99 |
ENST00000394863.3
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr19_+_16999654 | 0.99 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr12_-_29936731 | 0.98 |
ENST00000552618.1
ENST00000539277.1 ENST00000551659.1 |
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr22_-_24641027 | 0.97 |
ENST00000398292.3
ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5
|
gamma-glutamyltransferase 5 |
| chr1_-_59249732 | 0.97 |
ENST00000371222.2
|
JUN
|
jun proto-oncogene |
| chr16_-_87417351 | 0.97 |
ENST00000311635.7
|
FBXO31
|
F-box protein 31 |
| chr12_-_22094159 | 0.97 |
ENST00000538350.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr8_-_20161466 | 0.96 |
ENST00000381569.1
|
LZTS1
|
leucine zipper, putative tumor suppressor 1 |
| chr22_+_22764088 | 0.95 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr2_+_168725458 | 0.94 |
ENST00000392690.3
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr11_+_75273101 | 0.92 |
ENST00000533603.1
ENST00000358171.3 ENST00000526242.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr11_-_75379612 | 0.92 |
ENST00000526740.1
|
MAP6
|
microtubule-associated protein 6 |
| chr17_-_10017864 | 0.89 |
ENST00000323816.4
|
GAS7
|
growth arrest-specific 7 |
| chr10_-_75193308 | 0.87 |
ENST00000299432.2
|
MSS51
|
MSS51 mitochondrial translational activator |
| chr16_-_4987065 | 0.85 |
ENST00000590782.2
ENST00000345988.2 |
PPL
|
periplakin |
| chr12_+_7037461 | 0.83 |
ENST00000396684.2
|
ATN1
|
atrophin 1 |
| chrX_-_48328551 | 0.82 |
ENST00000376876.3
|
SLC38A5
|
solute carrier family 38, member 5 |
| chr4_+_39640787 | 0.82 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr11_-_75379479 | 0.80 |
ENST00000434603.2
|
MAP6
|
microtubule-associated protein 6 |
| chr19_+_13106383 | 0.79 |
ENST00000397661.2
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr12_-_108991812 | 0.78 |
ENST00000392806.3
ENST00000547567.1 |
TMEM119
|
transmembrane protein 119 |
| chr10_-_14372870 | 0.78 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr3_-_15374659 | 0.73 |
ENST00000426925.1
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr4_-_41216492 | 0.72 |
ENST00000503503.1
ENST00000509446.1 ENST00000503264.1 ENST00000508707.1 ENST00000508593.1 |
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr1_-_151119087 | 0.71 |
ENST00000341697.3
ENST00000368914.3 |
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chrX_-_48328631 | 0.71 |
ENST00000429543.1
ENST00000317669.5 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr4_-_819901 | 0.70 |
ENST00000304062.6
|
CPLX1
|
complexin 1 |
| chr1_-_235813290 | 0.69 |
ENST00000391854.2
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4 |
| chr12_+_101188718 | 0.68 |
ENST00000299222.9
ENST00000392977.3 |
ANO4
|
anoctamin 4 |
| chr5_+_140762268 | 0.68 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr15_-_88799661 | 0.67 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr6_+_39760129 | 0.66 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr15_-_88799384 | 0.65 |
ENST00000540489.2
ENST00000557856.1 ENST00000558676.1 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr12_+_47473369 | 0.65 |
ENST00000546455.1
|
PCED1B
|
PC-esterase domain containing 1B |
| chr3_-_195603566 | 0.65 |
ENST00000424563.1
ENST00000411741.1 |
TNK2
|
tyrosine kinase, non-receptor, 2 |
| chr4_-_41216619 | 0.65 |
ENST00000508676.1
ENST00000506352.1 ENST00000295974.8 |
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr19_-_46288917 | 0.64 |
ENST00000537879.1
ENST00000596586.1 ENST00000595946.1 |
DMWD
AC011530.4
|
dystrophia myotonica, WD repeat containing Uncharacterized protein |
| chr9_+_34990219 | 0.63 |
ENST00000541010.1
ENST00000454002.2 ENST00000545841.1 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr7_-_130418888 | 0.63 |
ENST00000310992.4
|
KLF14
|
Kruppel-like factor 14 |
| chr12_-_8765446 | 0.63 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr17_+_3627185 | 0.63 |
ENST00000325418.4
|
GSG2
|
germ cell associated 2 (haspin) |
| chr12_-_57410304 | 0.61 |
ENST00000441881.1
ENST00000458521.2 |
TAC3
|
tachykinin 3 |
| chr20_+_34680620 | 0.61 |
ENST00000430276.1
ENST00000373950.2 ENST00000452261.1 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr7_+_143080063 | 0.59 |
ENST00000446634.1
|
ZYX
|
zyxin |
| chr17_+_40440094 | 0.59 |
ENST00000546010.2
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chr6_+_39760783 | 0.57 |
ENST00000398904.2
ENST00000538976.1 |
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr20_-_56284816 | 0.57 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_-_41707778 | 0.57 |
ENST00000337495.5
ENST00000372597.1 ENST00000372596.1 |
SCMH1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr19_-_6481759 | 0.56 |
ENST00000588421.1
|
DENND1C
|
DENN/MADD domain containing 1C |
| chr7_+_149416439 | 0.56 |
ENST00000497895.1
|
KRBA1
|
KRAB-A domain containing 1 |
| chr19_-_42636543 | 0.56 |
ENST00000528894.4
ENST00000560804.2 ENST00000560558.1 ENST00000560398.1 ENST00000526816.2 |
POU2F2
|
POU class 2 homeobox 2 |
| chr20_-_36889127 | 0.56 |
ENST00000279024.4
|
KIAA1755
|
KIAA1755 |
| chr10_-_102989551 | 0.55 |
ENST00000370193.2
|
LBX1
|
ladybird homeobox 1 |
| chrX_+_153029633 | 0.55 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chr16_-_54320675 | 0.53 |
ENST00000329734.3
|
IRX3
|
iroquois homeobox 3 |
| chr19_+_50148087 | 0.53 |
ENST00000601038.1
ENST00000595242.1 |
SCAF1
|
SR-related CTD-associated factor 1 |
| chr13_-_80915059 | 0.53 |
ENST00000377104.3
|
SPRY2
|
sprouty homolog 2 (Drosophila) |
| chr4_-_41216473 | 0.51 |
ENST00000513140.1
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr11_-_1619524 | 0.51 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr1_+_185703513 | 0.48 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr11_-_33913708 | 0.48 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr2_-_73053126 | 0.48 |
ENST00000272427.6
ENST00000410104.1 |
EXOC6B
|
exocyst complex component 6B |
| chr20_+_42143053 | 0.48 |
ENST00000373135.3
ENST00000444063.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr12_+_122516626 | 0.47 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr3_+_3841108 | 0.46 |
ENST00000319331.3
|
LRRN1
|
leucine rich repeat neuronal 1 |
| chr11_-_101454658 | 0.45 |
ENST00000344327.3
|
TRPC6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr2_+_113033164 | 0.44 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr2_-_220173685 | 0.44 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chrX_-_151143140 | 0.44 |
ENST00000393914.3
ENST00000370328.3 ENST00000370325.1 |
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr2_+_27237615 | 0.43 |
ENST00000458529.1
ENST00000402218.1 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr19_-_49522727 | 0.43 |
ENST00000600007.1
|
CTB-60B18.10
|
CTB-60B18.10 |
| chrX_+_24167828 | 0.43 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr11_-_75380165 | 0.42 |
ENST00000304771.3
|
MAP6
|
microtubule-associated protein 6 |
| chr6_+_30852738 | 0.42 |
ENST00000508312.1
ENST00000512336.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_-_3859089 | 0.41 |
ENST00000396979.1
|
RHOG
|
ras homolog family member G |
| chr12_+_101188547 | 0.40 |
ENST00000546991.1
ENST00000392979.3 |
ANO4
|
anoctamin 4 |
| chr5_-_151066514 | 0.40 |
ENST00000538026.1
ENST00000522348.1 ENST00000521569.1 |
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr8_-_37411648 | 0.40 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr9_+_133569108 | 0.39 |
ENST00000372358.5
ENST00000546165.1 ENST00000372352.3 ENST00000372351.3 ENST00000372350.3 ENST00000495699.2 |
EXOSC2
|
exosome component 2 |
| chr19_-_42636617 | 0.38 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr11_+_74951948 | 0.38 |
ENST00000562197.2
|
TPBGL
|
trophoblast glycoprotein-like |
| chr3_+_123813543 | 0.38 |
ENST00000360013.3
|
KALRN
|
kalirin, RhoGEF kinase |
| chr7_-_69062391 | 0.37 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr1_+_226411319 | 0.37 |
ENST00000542034.1
ENST00000366810.5 |
MIXL1
|
Mix paired-like homeobox |
| chr7_-_100494222 | 0.37 |
ENST00000445236.1
|
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr13_-_88323514 | 0.37 |
ENST00000441617.1
|
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr9_-_73029540 | 0.37 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr10_+_99627889 | 0.36 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr18_+_20715416 | 0.35 |
ENST00000580153.1
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr10_+_102821551 | 0.35 |
ENST00000370200.5
|
KAZALD1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chr4_-_102268708 | 0.35 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr10_+_105314881 | 0.34 |
ENST00000437579.1
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr11_-_117166201 | 0.34 |
ENST00000510915.1
|
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr20_-_48532019 | 0.33 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr16_-_67997947 | 0.33 |
ENST00000537830.2
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr16_+_2286726 | 0.33 |
ENST00000382437.4
ENST00000569184.1 |
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr4_-_103749179 | 0.32 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_+_15783222 | 0.32 |
ENST00000359621.4
|
CELA2A
|
chymotrypsin-like elastase family, member 2A |
| chr20_+_34020827 | 0.31 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr4_-_185139062 | 0.31 |
ENST00000296741.2
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr4_-_103749205 | 0.31 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_+_102268904 | 0.30 |
ENST00000527564.1
ENST00000529296.1 |
AP001816.1
|
Uncharacterized protein |
| chr4_-_84030996 | 0.30 |
ENST00000411416.2
|
PLAC8
|
placenta-specific 8 |
| chr16_-_17564738 | 0.28 |
ENST00000261381.6
|
XYLT1
|
xylosyltransferase I |
| chr12_-_53207842 | 0.28 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr6_+_30689401 | 0.27 |
ENST00000396389.1
ENST00000396384.1 |
TUBB
|
tubulin, beta class I |
| chrX_-_30595959 | 0.27 |
ENST00000378962.3
|
CXorf21
|
chromosome X open reading frame 21 |
| chr12_+_3600356 | 0.26 |
ENST00000382622.3
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr6_+_30853002 | 0.26 |
ENST00000421124.2
ENST00000512725.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_-_71179699 | 0.25 |
ENST00000497355.1
|
FOXP1
|
forkhead box P1 |
| chr4_-_114682719 | 0.25 |
ENST00000394522.3
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr7_+_102004322 | 0.25 |
ENST00000496391.1
|
PRKRIP1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr11_-_18813353 | 0.24 |
ENST00000358540.2
ENST00000396171.4 ENST00000396167.2 |
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr4_-_114682597 | 0.24 |
ENST00000394524.3
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr17_+_30593195 | 0.24 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr21_-_47706098 | 0.24 |
ENST00000426537.1
|
MCM3AP
|
minichromosome maintenance complex component 3 associated protein |
| chr1_-_153508460 | 0.23 |
ENST00000462776.2
|
S100A6
|
S100 calcium binding protein A6 |
| chr12_-_56583243 | 0.23 |
ENST00000550164.1
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr19_-_57335682 | 0.22 |
ENST00000593931.1
|
ZIM2
|
zinc finger, imprinted 2 |
| chr5_+_133861339 | 0.22 |
ENST00000282605.4
ENST00000361895.2 ENST00000402835.1 |
PHF15
|
jade family PHD finger 2 |
| chr5_-_43412418 | 0.21 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr7_-_142724282 | 0.21 |
ENST00000350513.2
|
OR9A2
|
olfactory receptor, family 9, subfamily A, member 2 |
| chr16_+_24621546 | 0.21 |
ENST00000566108.1
|
CTD-2540M10.1
|
CTD-2540M10.1 |
| chr8_+_75015699 | 0.19 |
ENST00000522498.1
|
RP11-6I2.4
|
RP11-6I2.4 |
| chr11_-_74204692 | 0.19 |
ENST00000528085.1
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr20_+_44098346 | 0.19 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr11_-_34535297 | 0.19 |
ENST00000532417.1
|
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr12_-_47473642 | 0.19 |
ENST00000266581.4
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr12_-_47473557 | 0.18 |
ENST00000321382.3
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr1_-_149783914 | 0.18 |
ENST00000369167.1
ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF
|
histone cluster 2, H2bf |
| chr12_+_68042517 | 0.18 |
ENST00000393555.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr3_+_40547483 | 0.18 |
ENST00000420891.1
ENST00000314529.6 ENST00000418905.1 |
ZNF620
|
zinc finger protein 620 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 1.5 | 4.6 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.5 | 6.0 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 1.3 | 3.9 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.8 | 2.4 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.6 | 1.9 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.5 | 1.1 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.4 | 1.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.4 | 1.2 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.4 | 2.7 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.4 | 1.5 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.4 | 2.5 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 | 1.3 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.3 | 1.2 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.3 | 1.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 1.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.3 | 1.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.2 | 2.2 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.2 | 1.8 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.2 | 0.4 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.2 | 2.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.6 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 | 0.6 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.2 | 5.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.0 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.2 | 1.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.4 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.5 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.1 | 0.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.9 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.5 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 1.5 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 1.5 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 0.9 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.3 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.6 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.3 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.1 | 1.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.4 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.9 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 2.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.4 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.4 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.5 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.1 | 1.0 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 8.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 4.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.8 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.4 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 3.5 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.6 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 1.0 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 2.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 1.8 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 1.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.9 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.4 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 1.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.8 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.8 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 4.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.9 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 2.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 7.7 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 2.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 6.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 3.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 5.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 3.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.6 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.0 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.3 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.1 | 0.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.3 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 1.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 2.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 2.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 1.0 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 4.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 2.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.1 | 4.6 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.7 | 2.7 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.5 | 2.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 6.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.3 | 0.9 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.3 | 1.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.3 | 1.9 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 2.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 1.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 7.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 0.5 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.2 | 3.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 1.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) neurotrophin receptor activity(GO:0005030) |
| 0.1 | 3.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.5 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 2.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 2.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 1.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.6 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 10.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.9 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 6.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 5.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 3.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 9.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 4.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 9.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 2.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 3.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |