Project
Illumina Body Map 2
Navigation
Downloads
Results for SOX13_SOX12
Z-value: 1.34
Transcription factors associated with SOX13_SOX12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
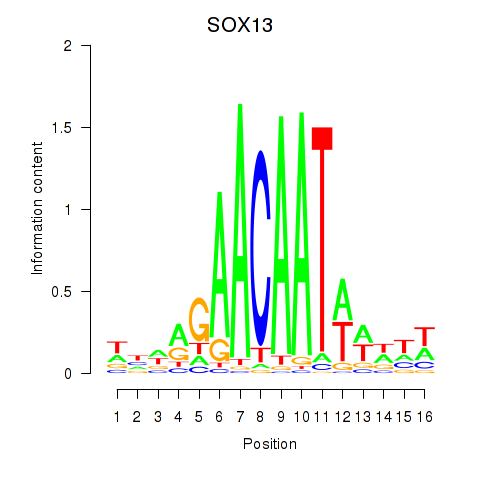
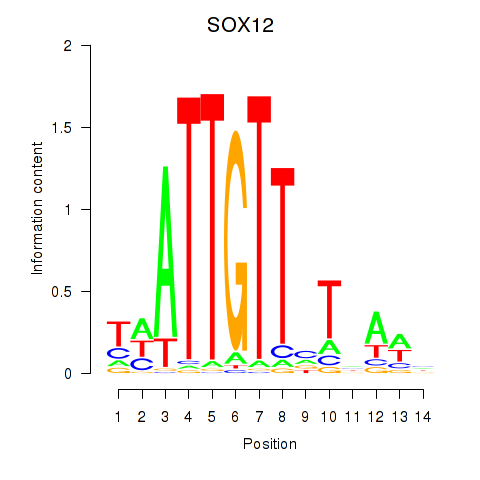
SOX13
|
ENSG00000143842.10 | SRY-box transcription factor 13 |
|
SOX12
|
ENSG00000177732.6 | SRY-box transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX12 | hg19_v2_chr20_+_306177_306211 | 0.18 | 3.3e-01 | Click! |
| SOX13 | hg19_v2_chr1_+_204042723_204042784 | 0.11 | 5.3e-01 | Click! |
Activity profile of SOX13_SOX12 motif
Sorted Z-values of SOX13_SOX12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_10573149 | 5.01 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr6_-_49712091 | 4.39 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr6_-_49712123 | 4.22 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr6_-_49712147 | 3.96 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr6_-_49712072 | 3.80 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr12_-_11463353 | 3.42 |
ENST00000279575.1
ENST00000535904.1 ENST00000445719.2 |
PRB4
|
proline-rich protein BstNI subfamily 4 |
| chr22_-_50523760 | 3.18 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr4_+_36283213 | 3.05 |
ENST00000357504.3
|
DTHD1
|
death domain containing 1 |
| chr20_-_52612468 | 2.98 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr19_+_55105085 | 2.92 |
ENST00000251372.3
ENST00000453777.1 |
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1 |
| chr22_-_50523843 | 2.85 |
ENST00000535444.1
ENST00000431262.2 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr22_-_50523807 | 2.84 |
ENST00000442311.1
ENST00000538737.1 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr12_-_10588539 | 2.71 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr20_-_52612705 | 2.69 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr12_+_11081828 | 2.55 |
ENST00000381847.3
ENST00000396400.3 |
PRH2
|
proline-rich protein HaeIII subfamily 2 |
| chr14_+_21423611 | 2.46 |
ENST00000304625.2
|
RNASE2
|
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) |
| chr4_-_57547454 | 2.44 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr22_-_50523688 | 2.40 |
ENST00000450140.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr12_-_11508520 | 2.37 |
ENST00000545626.1
ENST00000500254.2 |
PRB1
|
proline-rich protein BstNI subfamily 1 |
| chr19_-_14785698 | 2.34 |
ENST00000344373.4
ENST00000595472.1 |
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr19_-_14785674 | 2.33 |
ENST00000253673.5
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr7_-_38305279 | 2.31 |
ENST00000443402.2
|
TRGC1
|
T cell receptor gamma constant 1 |
| chr19_-_14785622 | 2.29 |
ENST00000443157.2
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr10_-_28270795 | 2.25 |
ENST00000545014.1
|
ARMC4
|
armadillo repeat containing 4 |
| chr4_-_74864386 | 2.24 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr1_-_153363452 | 2.23 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr17_+_61151306 | 2.17 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr12_-_10601963 | 2.17 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr12_+_8666126 | 2.12 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr1_+_66797687 | 2.09 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr12_-_11548496 | 2.07 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr2_+_169923504 | 2.07 |
ENST00000357546.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr2_+_169923577 | 2.06 |
ENST00000432060.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr1_-_162838551 | 2.06 |
ENST00000367910.1
ENST00000367912.2 ENST00000367911.2 |
C1orf110
|
chromosome 1 open reading frame 110 |
| chr11_+_60223225 | 2.04 |
ENST00000524807.1
ENST00000345732.4 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr7_+_142364193 | 2.02 |
ENST00000390397.2
|
TRBV24-1
|
T cell receptor beta variable 24-1 |
| chr11_+_60223312 | 2.02 |
ENST00000532491.1
ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr15_-_41166414 | 1.96 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr18_-_24445664 | 1.91 |
ENST00000578776.1
|
AQP4
|
aquaporin 4 |
| chr2_+_102927962 | 1.85 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr14_+_22180536 | 1.79 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr8_+_24151553 | 1.76 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr7_-_142198049 | 1.76 |
ENST00000471935.1
|
TRBV11-2
|
T cell receptor beta variable 11-2 |
| chr1_+_77333117 | 1.75 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr12_-_45315625 | 1.71 |
ENST00000552993.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr15_-_79383102 | 1.71 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr4_-_90759440 | 1.70 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr12_-_10324716 | 1.68 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr15_+_51669513 | 1.68 |
ENST00000558426.1
|
GLDN
|
gliomedin |
| chr5_+_54320078 | 1.66 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr12_+_60058458 | 1.63 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr12_-_11422630 | 1.62 |
ENST00000381842.3
ENST00000538488.1 |
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr2_+_90153696 | 1.61 |
ENST00000417279.2
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr6_-_133055815 | 1.61 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr7_-_30008849 | 1.58 |
ENST00000409497.1
|
SCRN1
|
secernin 1 |
| chr14_+_22089953 | 1.53 |
ENST00000542354.1
|
TRAV1-1
|
T cell receptor alpha variable 1-1 |
| chrX_+_107288280 | 1.50 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrY_+_15016013 | 1.47 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr2_-_89327228 | 1.46 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr5_+_35852797 | 1.46 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr2_-_89385283 | 1.42 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr6_-_133055896 | 1.41 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr14_+_22580233 | 1.41 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr6_-_41254403 | 1.41 |
ENST00000589614.1
ENST00000334475.6 ENST00000591620.1 ENST00000244709.4 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr12_-_10605929 | 1.40 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr12_-_11422739 | 1.40 |
ENST00000279573.7
|
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr6_-_32498046 | 1.39 |
ENST00000374975.3
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5 |
| chr11_+_128563652 | 1.39 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_128222103 | 1.38 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr19_-_54604083 | 1.38 |
ENST00000391761.1
ENST00000356532.3 ENST00000359649.4 ENST00000358375.4 ENST00000391760.1 ENST00000351806.4 |
OSCAR
|
osteoclast associated, immunoglobulin-like receptor |
| chr2_+_90198535 | 1.35 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr1_-_68915610 | 1.35 |
ENST00000262340.5
|
RPE65
|
retinal pigment epithelium-specific protein 65kDa |
| chrX_+_107288197 | 1.35 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr14_+_22217447 | 1.34 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr12_-_10282836 | 1.34 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr10_-_10836919 | 1.33 |
ENST00000602763.1
ENST00000415590.2 ENST00000434919.2 |
SFTA1P
|
surfactant associated 1, pseudogene |
| chr7_-_115608304 | 1.33 |
ENST00000457268.1
|
TFEC
|
transcription factor EC |
| chr12_+_47610315 | 1.32 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr5_+_142286887 | 1.32 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr4_+_102734967 | 1.31 |
ENST00000444316.2
|
BANK1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr1_+_115642293 | 1.29 |
ENST00000448680.1
|
RP4-666F24.3
|
RP4-666F24.3 |
| chr17_+_56315787 | 1.29 |
ENST00000262290.4
ENST00000421678.2 |
LPO
|
lactoperoxidase |
| chr4_-_108204904 | 1.28 |
ENST00000510463.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr3_+_111717511 | 1.26 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr14_+_22739823 | 1.24 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr1_+_87012753 | 1.24 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr20_+_31870927 | 1.24 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr17_+_61271355 | 1.22 |
ENST00000583356.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr17_+_12453278 | 1.21 |
ENST00000577679.1
|
LINC00670
|
long intergenic non-protein coding RNA 670 |
| chr4_-_70518941 | 1.21 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr12_-_85430024 | 1.21 |
ENST00000547836.1
ENST00000532498.2 |
TSPAN19
|
tetraspanin 19 |
| chr4_-_108204846 | 1.20 |
ENST00000513208.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr11_-_85430356 | 1.19 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr3_+_32433154 | 1.18 |
ENST00000334983.5
ENST00000349718.4 |
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr14_-_52436247 | 1.17 |
ENST00000597846.1
|
AL358333.1
|
HCG2013195; Uncharacterized protein |
| chr12_-_45307711 | 1.15 |
ENST00000333837.4
ENST00000551949.1 |
NELL2
|
NEL-like 2 (chicken) |
| chrX_+_102192200 | 1.15 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr1_-_157670528 | 1.14 |
ENST00000368186.5
ENST00000496769.1 |
FCRL3
|
Fc receptor-like 3 |
| chr4_-_74853897 | 1.13 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr2_+_90273679 | 1.13 |
ENST00000423080.2
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr11_-_104893863 | 1.13 |
ENST00000260315.3
ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5
|
caspase 5, apoptosis-related cysteine peptidase |
| chr12_+_25205568 | 1.12 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr3_+_111717600 | 1.12 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr1_+_161677034 | 1.12 |
ENST00000349527.4
ENST00000309691.6 ENST00000294796.4 ENST00000367953.3 ENST00000367950.1 |
FCRLA
|
Fc receptor-like A |
| chr11_-_102595512 | 1.11 |
ENST00000438475.2
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr11_+_4470525 | 1.11 |
ENST00000325719.4
|
OR52K2
|
olfactory receptor, family 52, subfamily K, member 2 |
| chr11_-_85430088 | 1.11 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr8_+_24151620 | 1.10 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr18_-_24445729 | 1.09 |
ENST00000383168.4
|
AQP4
|
aquaporin 4 |
| chr11_-_85430204 | 1.09 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr2_-_69180012 | 1.09 |
ENST00000481498.1
|
GKN2
|
gastrokine 2 |
| chr12_-_11036844 | 1.09 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr12_-_81992111 | 1.08 |
ENST00000443686.3
ENST00000407050.4 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr3_+_130279178 | 1.07 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr2_-_157198860 | 1.06 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr19_+_35609380 | 1.06 |
ENST00000604621.1
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr1_+_66458072 | 1.05 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr12_+_25205628 | 1.05 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr12_-_11002063 | 1.04 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr2_-_216878305 | 1.04 |
ENST00000263268.6
|
MREG
|
melanoregulin |
| chr2_-_130902567 | 1.04 |
ENST00000457413.1
ENST00000392984.3 ENST00000409128.1 ENST00000441670.1 ENST00000409943.3 ENST00000409234.3 ENST00000310463.6 |
CCDC74B
|
coiled-coil domain containing 74B |
| chr11_-_85430163 | 1.03 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr1_-_158656488 | 1.03 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr7_-_38289173 | 1.03 |
ENST00000436911.2
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr15_-_80263506 | 1.03 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr3_-_167191814 | 1.02 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr11_-_102595681 | 1.01 |
ENST00000236826.3
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr2_+_204571375 | 1.01 |
ENST00000374478.4
|
CD28
|
CD28 molecule |
| chr8_-_108510224 | 1.00 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr6_-_131299929 | 0.99 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr14_+_22748980 | 0.99 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr10_+_49917751 | 0.98 |
ENST00000325239.5
ENST00000413659.2 |
WDFY4
|
WDFY family member 4 |
| chr8_-_116504448 | 0.97 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr11_+_122709200 | 0.95 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr4_-_48116540 | 0.95 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr21_+_38593701 | 0.94 |
ENST00000440629.1
|
AP001432.14
|
AP001432.14 |
| chr1_+_161676739 | 0.94 |
ENST00000236938.6
ENST00000367959.2 ENST00000546024.1 ENST00000540521.1 ENST00000367949.2 ENST00000350710.3 ENST00000540926.1 |
FCRLA
|
Fc receptor-like A |
| chr3_+_32433363 | 0.93 |
ENST00000465248.1
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr2_+_90211643 | 0.93 |
ENST00000390277.2
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr19_-_22018966 | 0.92 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr14_+_21387491 | 0.92 |
ENST00000258817.2
|
RP11-84C10.2
|
RP11-84C10.2 |
| chrX_+_36254051 | 0.92 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr3_-_181160240 | 0.91 |
ENST00000460993.1
|
RP11-275H4.1
|
RP11-275H4.1 |
| chr2_-_89278535 | 0.91 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr2_+_196440692 | 0.90 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr6_-_19804973 | 0.90 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr8_+_104831554 | 0.90 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chrX_+_107288239 | 0.90 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr11_+_35201826 | 0.90 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_81106951 | 0.89 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr4_-_112993808 | 0.89 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr12_-_68835991 | 0.88 |
ENST00000546086.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr11_+_36397731 | 0.88 |
ENST00000524380.1
|
PRR5L
|
proline rich 5 like |
| chr11_-_102651343 | 0.88 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr6_-_32557610 | 0.88 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr11_+_128563948 | 0.88 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_+_71337834 | 0.87 |
ENST00000304887.5
|
MUC7
|
mucin 7, secreted |
| chr1_+_87012922 | 0.86 |
ENST00000263723.5
|
CLCA4
|
chloride channel accessory 4 |
| chr12_-_10562356 | 0.86 |
ENST00000309384.1
|
KLRC4
|
killer cell lectin-like receptor subfamily C, member 4 |
| chr8_+_32579321 | 0.86 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr12_+_25205666 | 0.86 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr14_+_23012122 | 0.85 |
ENST00000390534.1
|
TRAJ3
|
T cell receptor alpha joining 3 |
| chr2_-_99279928 | 0.85 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr12_-_11184006 | 0.85 |
ENST00000390675.2
|
TAS2R31
|
taste receptor, type 2, member 31 |
| chr17_-_47286244 | 0.84 |
ENST00000503070.1
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr12_+_8995832 | 0.84 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr11_+_60467047 | 0.84 |
ENST00000300226.2
|
MS4A8
|
membrane-spanning 4-domains, subfamily A, member 8 |
| chr17_+_56315936 | 0.84 |
ENST00000543544.1
|
LPO
|
lactoperoxidase |
| chr8_+_32579271 | 0.84 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr8_+_104831440 | 0.83 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_-_243418621 | 0.83 |
ENST00000366544.1
ENST00000366543.1 |
CEP170
|
centrosomal protein 170kDa |
| chr12_-_10607084 | 0.83 |
ENST00000408006.3
ENST00000544822.1 ENST00000536188.1 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr10_-_56561022 | 0.82 |
ENST00000373965.2
ENST00000414778.1 ENST00000395438.1 ENST00000409834.1 ENST00000395445.1 ENST00000395446.1 ENST00000395442.1 ENST00000395440.1 ENST00000395432.2 ENST00000361849.3 ENST00000395433.1 ENST00000320301.6 ENST00000395430.1 ENST00000437009.1 |
PCDH15
|
protocadherin-related 15 |
| chr14_+_56584414 | 0.82 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr7_-_102236184 | 0.81 |
ENST00000521397.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr11_+_5711010 | 0.81 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr6_+_106534192 | 0.80 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr19_+_55141948 | 0.78 |
ENST00000396332.4
ENST00000427581.2 |
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
| chr7_-_36764004 | 0.78 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr4_+_174818390 | 0.78 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr9_-_28670283 | 0.78 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr16_+_84178850 | 0.78 |
ENST00000334315.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr21_+_43619796 | 0.78 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr2_+_158114051 | 0.78 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr7_-_36764062 | 0.77 |
ENST00000435386.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr6_+_32605195 | 0.76 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr8_-_67940755 | 0.76 |
ENST00000324682.5
|
PPP1R42
|
protein phosphatase 1, regulatory subunit 42 |
| chr4_+_165675197 | 0.76 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr6_-_88001706 | 0.76 |
ENST00000369576.2
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr4_-_185458660 | 0.76 |
ENST00000605270.1
|
RP11-326I11.4
|
RP11-326I11.4 |
| chr2_-_158300556 | 0.75 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr19_-_9003586 | 0.75 |
ENST00000380951.5
|
MUC16
|
mucin 16, cell surface associated |
| chr4_+_159131630 | 0.75 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr3_+_141596371 | 0.75 |
ENST00000495216.1
|
ATP1B3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr7_+_64126503 | 0.75 |
ENST00000360117.3
|
ZNF107
|
zinc finger protein 107 |
| chr2_-_25100893 | 0.74 |
ENST00000433852.1
|
ADCY3
|
adenylate cyclase 3 |
| chrY_-_21738068 | 0.74 |
ENST00000599485.1
|
AC009977.1
|
Uncharacterized protein |
| chr4_-_38858428 | 0.73 |
ENST00000436693.2
ENST00000508254.1 ENST00000514655.1 ENST00000506146.1 |
TLR6
TLR1
|
toll-like receptor 6 toll-like receptor 1 |
| chr1_+_158815588 | 0.73 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chrX_-_19688475 | 0.73 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX13_SOX12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0018969 | thiocyanate metabolic process(GO:0018969) |
| 0.7 | 2.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.6 | 1.2 | GO:0009593 | detection of chemical stimulus(GO:0009593) |
| 0.5 | 1.5 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.5 | 11.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.4 | 1.4 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.4 | 4.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 1.4 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.3 | 1.7 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.3 | 4.9 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.3 | 0.9 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.3 | 4.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 1.3 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.3 | 0.8 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.3 | 1.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.2 | 0.7 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.2 | 0.7 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.2 | 0.7 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.2 | 1.4 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.2 | 0.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 2.1 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.2 | 1.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 2.7 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 0.6 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.2 | 1.6 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 1.0 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.2 | 0.8 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.2 | 0.7 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.2 | 0.9 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.2 | 0.7 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 0.5 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 1.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.2 | 0.5 | GO:1904346 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.2 | 0.9 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.2 | 0.3 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 1.8 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 2.9 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.2 | 1.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 2.6 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.2 | 2.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 3.0 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.2 | 3.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 | 1.7 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.7 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.3 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 0.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.3 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 4.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.9 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 1.0 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 2.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.3 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.1 | 0.3 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.7 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.6 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 1.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.3 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.1 | 0.9 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.3 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.1 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.5 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.3 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.3 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.1 | 0.4 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 9.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 2.2 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 2.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.9 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.1 | 0.4 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.7 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.3 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.1 | 2.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 0.2 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 1.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.7 | GO:0045345 | positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 | 1.9 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.5 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.1 | 2.3 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 0.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 2.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.2 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 1.7 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.6 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 0.3 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 1.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 5.4 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 1.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.4 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.5 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.7 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.1 | 1.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.4 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.2 | GO:0052416 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.1 | 0.6 | GO:0050713 | negative regulation of interleukin-1 secretion(GO:0050711) negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.7 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.3 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 10.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.6 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.6 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.9 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.2 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 0.1 | 0.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 1.8 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.5 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.2 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.3 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.2 | GO:1903525 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.5 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 1.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 4.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.3 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0060928 | cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 | 0.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 31.3 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.4 | GO:0071287 | cellular response to manganese ion(GO:0071287) |
| 0.0 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.1 | GO:1903979 | regulation of microglial cell activation(GO:1903978) negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 0.4 | GO:0002252 | immune effector process(GO:0002252) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.0 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.5 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.5 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0032916 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 0.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 4.7 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 2.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.2 | GO:1901660 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 1.3 | GO:0050871 | positive regulation of B cell activation(GO:0050871) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.2 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 2.8 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.7 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 1.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.8 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.9 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 1.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.3 | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.7 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.2 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.8 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.6 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 1.6 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.0 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.3 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.6 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.4 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.2 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.0 | GO:0006174 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.5 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.3 | 0.9 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.2 | 2.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 0.7 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.2 | 21.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.2 | 3.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.3 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 2.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.3 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.8 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.5 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.6 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 3.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 11.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.7 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 3.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.3 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 4.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.2 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 6.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 1.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 5.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 14.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 6.9 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 1.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.7 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 6.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.8 | 2.4 | GO:0036393 | thiocyanate peroxidase activity(GO:0036393) |
| 0.6 | 3.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.5 | 1.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 1.8 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.4 | 1.6 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.4 | 1.5 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.4 | 5.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.3 | 1.4 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.3 | 1.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 2.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.3 | 0.6 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.3 | 1.9 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.3 | 2.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 0.8 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.2 | 0.7 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.2 | 0.7 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 1.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 1.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.2 | 3.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 2.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 5.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 2.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 0.5 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.2 | 0.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 5.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 2.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 2.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.4 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.1 | 0.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.6 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 1.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.8 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.3 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.4 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 4.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.7 | GO:0005124 | N-formyl peptide receptor activity(GO:0004982) scavenger receptor binding(GO:0005124) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 2.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.3 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.6 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 17.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.2 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 2.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.1 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 0.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.2 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.1 | 0.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.2 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 9.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 2.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.5 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.2 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.4 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 2.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 4.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.5 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0015149 | hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.0 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 3.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 2.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 5.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 2.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 5.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 3.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 10.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 2.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 8.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 4.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 5.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.2 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 2.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 2.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 2.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.8 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |