Project
Illumina Body Map 2
Navigation
Downloads
Results for SOX30
Z-value: 0.88
Transcription factors associated with SOX30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX30
|
ENSG00000039600.6 | SRY-box transcription factor 30 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX30 | hg19_v2_chr5_-_157079428_157079452 | 0.14 | 4.5e-01 | Click! |
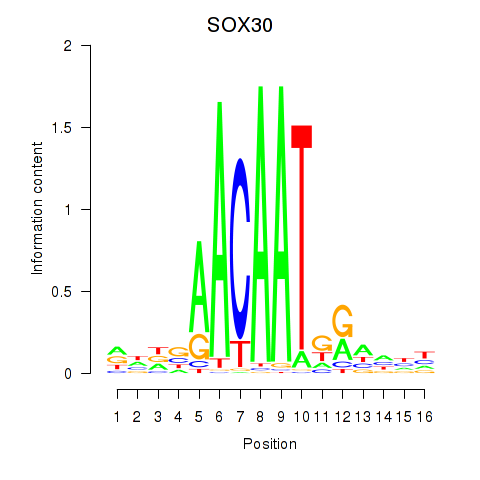
Activity profile of SOX30 motif
Sorted Z-values of SOX30 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_11548496 | 3.91 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr12_-_11463353 | 3.68 |
ENST00000279575.1
ENST00000535904.1 ENST00000445719.2 |
PRB4
|
proline-rich protein BstNI subfamily 4 |
| chr12_-_11508520 | 3.38 |
ENST00000545626.1
ENST00000500254.2 |
PRB1
|
proline-rich protein BstNI subfamily 1 |
| chr12_+_11081828 | 3.10 |
ENST00000381847.3
ENST00000396400.3 |
PRH2
|
proline-rich protein HaeIII subfamily 2 |
| chr12_-_11002063 | 3.00 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr12_-_11036844 | 2.55 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr12_-_11422630 | 2.49 |
ENST00000381842.3
ENST00000538488.1 |
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr12_-_11422739 | 2.04 |
ENST00000279573.7
|
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chrY_+_15016013 | 2.01 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr3_+_181429704 | 1.62 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr10_-_28270795 | 1.48 |
ENST00000545014.1
|
ARMC4
|
armadillo repeat containing 4 |
| chr4_+_77356248 | 1.40 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr2_-_158182410 | 1.35 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr5_-_82969405 | 1.30 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr7_+_100770328 | 1.20 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr3_-_165555200 | 1.05 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr7_-_121944491 | 1.00 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr8_-_70016408 | 0.97 |
ENST00000518540.1
|
RP11-600K15.1
|
RP11-600K15.1 |
| chr5_+_139175380 | 0.93 |
ENST00000274710.3
|
PSD2
|
pleckstrin and Sec7 domain containing 2 |
| chr3_-_19988462 | 0.93 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr1_+_47489240 | 0.88 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr6_+_12290586 | 0.87 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr11_+_63057412 | 0.84 |
ENST00000544661.1
|
SLC22A10
|
solute carrier family 22, member 10 |
| chr16_+_87636474 | 0.84 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chrX_-_24690771 | 0.82 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr5_-_82969363 | 0.81 |
ENST00000503117.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr2_-_158182322 | 0.78 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr14_+_100070869 | 0.77 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr1_-_204463829 | 0.75 |
ENST00000429009.1
ENST00000415899.1 |
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr1_-_26231589 | 0.74 |
ENST00000374291.1
|
STMN1
|
stathmin 1 |
| chr17_-_38821373 | 0.68 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr6_+_126240442 | 0.68 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr3_-_167191814 | 0.66 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr1_-_201476274 | 0.64 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr2_+_179184955 | 0.63 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chrX_+_114874727 | 0.63 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr2_-_166702601 | 0.60 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr8_+_24151553 | 0.59 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr1_-_201476220 | 0.59 |
ENST00000526723.1
ENST00000524951.1 |
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr12_-_10251576 | 0.58 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr13_+_98795664 | 0.58 |
ENST00000376581.5
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr12_-_10251539 | 0.58 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr12_-_10251603 | 0.57 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr12_-_10588539 | 0.44 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr9_+_103235365 | 0.44 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr4_-_70725856 | 0.44 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr12_-_66317967 | 0.43 |
ENST00000601398.1
|
AC090673.2
|
Uncharacterized protein |
| chr5_+_140729649 | 0.41 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr1_+_185703513 | 0.41 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr13_+_110958124 | 0.39 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr4_-_176733897 | 0.38 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr5_+_140739537 | 0.38 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr5_+_170288856 | 0.38 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr10_-_62060232 | 0.38 |
ENST00000503925.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr7_-_82792215 | 0.37 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr5_+_140772381 | 0.36 |
ENST00000398604.2
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr5_+_140235469 | 0.36 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr11_-_85430356 | 0.35 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr5_+_38148582 | 0.35 |
ENST00000508853.1
|
CTD-2207A17.1
|
CTD-2207A17.1 |
| chr15_+_69857515 | 0.35 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr1_-_243418621 | 0.34 |
ENST00000366544.1
ENST00000366543.1 |
CEP170
|
centrosomal protein 170kDa |
| chr1_-_85930246 | 0.33 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr10_-_124459284 | 0.33 |
ENST00000432000.1
ENST00000329446.4 |
C10orf120
|
chromosome 10 open reading frame 120 |
| chr11_-_85430163 | 0.31 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr6_-_49712123 | 0.31 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr6_-_11779174 | 0.30 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr8_+_24151620 | 0.30 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr4_-_87028478 | 0.30 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr8_-_49533521 | 0.28 |
ENST00000523038.1
|
RP11-567J20.1
|
RP11-567J20.1 |
| chr11_-_123756334 | 0.28 |
ENST00000528595.1
ENST00000375026.2 |
TMEM225
|
transmembrane protein 225 |
| chr1_+_240255166 | 0.27 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr11_-_85430204 | 0.26 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chrX_+_102192200 | 0.25 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr8_-_82024290 | 0.25 |
ENST00000220597.4
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr1_+_228337553 | 0.25 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr12_+_80750134 | 0.25 |
ENST00000546620.1
|
OTOGL
|
otogelin-like |
| chr12_+_27623565 | 0.25 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr9_-_74383302 | 0.23 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr2_+_143635067 | 0.23 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr1_-_243418344 | 0.23 |
ENST00000366542.1
|
CEP170
|
centrosomal protein 170kDa |
| chr20_-_5426332 | 0.23 |
ENST00000420529.1
|
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr6_+_22146851 | 0.22 |
ENST00000606197.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr16_+_20499024 | 0.22 |
ENST00000593357.1
|
AC137056.1
|
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chr3_-_57583130 | 0.21 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr5_-_160279207 | 0.20 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr6_-_11779014 | 0.20 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr1_-_167883353 | 0.19 |
ENST00000545172.1
|
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr12_-_99288986 | 0.19 |
ENST00000552407.1
ENST00000551613.1 ENST00000548447.1 ENST00000546364.3 ENST00000552748.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_+_35415719 | 0.19 |
ENST00000474362.1
ENST00000374721.3 |
CREM
|
cAMP responsive element modulator |
| chr10_+_35415978 | 0.19 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr13_-_25754077 | 0.18 |
ENST00000413501.1
|
AMER2-AS1
|
AMER2 antisense RNA 1 (head to head) |
| chr1_-_167883327 | 0.17 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr1_-_198906528 | 0.17 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr1_-_200379180 | 0.16 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr3_+_19988566 | 0.16 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr20_-_62610982 | 0.16 |
ENST00000369886.3
ENST00000450107.1 |
SAMD10
|
sterile alpha motif domain containing 10 |
| chr20_+_30555805 | 0.16 |
ENST00000562532.2
|
XKR7
|
XK, Kell blood group complex subunit-related family, member 7 |
| chr11_-_85430088 | 0.15 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr6_-_89927151 | 0.14 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr11_+_128563652 | 0.14 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr10_+_35416090 | 0.14 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr5_-_146461027 | 0.13 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_-_200379129 | 0.13 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr2_-_169769787 | 0.12 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr10_+_35416223 | 0.11 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr1_-_243418650 | 0.10 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170kDa |
| chr1_+_74701062 | 0.10 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr14_-_90420862 | 0.09 |
ENST00000556005.1
ENST00000555872.1 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr10_-_61122934 | 0.08 |
ENST00000512919.1
|
FAM13C
|
family with sequence similarity 13, member C |
| chr7_-_142176790 | 0.08 |
ENST00000390369.2
|
TRBV7-4
|
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr11_+_63137251 | 0.07 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr15_+_93426514 | 0.07 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr4_+_17516788 | 0.07 |
ENST00000511148.2
|
CLRN2
|
clarin 2 |
| chr5_-_141338627 | 0.07 |
ENST00000231484.3
|
PCDH12
|
protocadherin 12 |
| chr4_+_71457970 | 0.07 |
ENST00000322937.6
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr4_-_186570679 | 0.07 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_+_19988736 | 0.07 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr12_+_93096759 | 0.07 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr17_+_7323634 | 0.06 |
ENST00000323675.3
|
SPEM1
|
spermatid maturation 1 |
| chr15_+_76629064 | 0.06 |
ENST00000290759.4
|
ISL2
|
ISL LIM homeobox 2 |
| chr14_+_101299520 | 0.06 |
ENST00000455531.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr12_-_49581152 | 0.06 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr22_-_30970560 | 0.06 |
ENST00000402369.1
ENST00000406361.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr6_-_134639180 | 0.06 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_+_71458012 | 0.05 |
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr14_+_36295504 | 0.04 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr14_-_90421028 | 0.03 |
ENST00000267544.9
ENST00000316738.7 ENST00000538485.2 ENST00000556609.1 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr6_-_134639235 | 0.02 |
ENST00000533224.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr6_-_161085291 | 0.02 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr1_+_244998602 | 0.02 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chrX_-_117107542 | 0.02 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr17_-_73149921 | 0.02 |
ENST00000481647.1
ENST00000470924.1 |
HN1
|
hematological and neurological expressed 1 |
| chr1_+_244998918 | 0.01 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr1_+_149239529 | 0.00 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX30
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.4 | 1.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.2 | 0.9 | GO:0060584 | nitric oxide transport(GO:0030185) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 1.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 1.6 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.4 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 1.5 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 0.3 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 1.6 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 1.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.8 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.4 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 4.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 3.0 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 1.2 | GO:0070527 | platelet aggregation(GO:0070527) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 2.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 1.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.2 | 0.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.7 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 2.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.8 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.7 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 4.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |