Project
Illumina Body Map 2
Navigation
Downloads
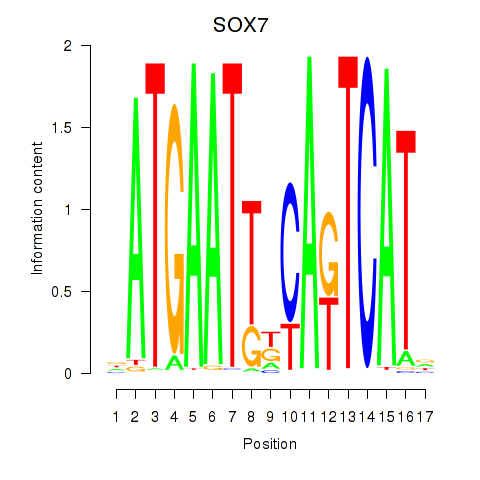
Results for SOX7
Z-value: 0.89
Transcription factors associated with SOX7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX7
|
ENSG00000171056.6 | SRY-box transcription factor 7 |
|
SOX7
|
ENSG00000258724.1 | SRY-box transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX7 | hg19_v2_chr8_-_10697281_10697365 | -0.39 | 2.8e-02 | Click! |
Activity profile of SOX7 motif
Sorted Z-values of SOX7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_20364030 | 7.19 |
ENST00000396134.2
ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD
|
uromodulin |
| chr16_-_20364122 | 6.29 |
ENST00000396138.4
ENST00000577168.1 |
UMOD
|
uromodulin |
| chr1_-_26394114 | 4.68 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr4_-_177190364 | 3.35 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr6_-_46048116 | 2.14 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr10_-_61495760 | 2.14 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr4_+_76995855 | 1.73 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chr1_-_186430222 | 1.58 |
ENST00000391997.2
|
PDC
|
phosducin |
| chr1_-_43751276 | 1.43 |
ENST00000423420.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr6_+_150690089 | 1.36 |
ENST00000392256.2
|
IYD
|
iodotyrosine deiodinase |
| chr10_+_5005445 | 1.35 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr6_+_150690028 | 1.32 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr3_+_167584414 | 1.29 |
ENST00000475324.1
|
RP11-298O21.2
|
RP11-298O21.2 |
| chr8_+_12803176 | 1.23 |
ENST00000524591.2
|
KIAA1456
|
KIAA1456 |
| chr6_+_140175987 | 1.21 |
ENST00000414038.1
ENST00000431609.1 |
RP5-899B16.1
|
RP5-899B16.1 |
| chr12_-_58027138 | 1.16 |
ENST00000341156.4
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr14_+_95048217 | 1.14 |
ENST00000557598.1
ENST00000556064.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr4_-_47983519 | 1.10 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr5_-_95158375 | 1.09 |
ENST00000512469.2
ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX
|
glutaredoxin (thioltransferase) |
| chr17_-_28661065 | 1.03 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr1_-_43751230 | 0.98 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr19_+_37837218 | 0.96 |
ENST00000591134.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr5_-_95158644 | 0.91 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr10_+_24738355 | 0.77 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr10_-_5046042 | 0.75 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr9_-_70465758 | 0.73 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr6_+_150690133 | 0.67 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
| chr12_-_56693758 | 0.66 |
ENST00000547298.1
ENST00000551936.1 ENST00000551253.1 ENST00000551473.1 |
CS
|
citrate synthase |
| chr19_-_22379753 | 0.64 |
ENST00000397121.2
|
ZNF676
|
zinc finger protein 676 |
| chr14_-_102771462 | 0.63 |
ENST00000522874.1
|
MOK
|
MOK protein kinase |
| chr14_+_50291993 | 0.57 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr17_-_66951382 | 0.56 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr19_-_5784610 | 0.55 |
ENST00000390672.2
ENST00000419421.2 |
PRR22
|
proline rich 22 |
| chr19_+_37837185 | 0.53 |
ENST00000541583.2
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr15_+_25068773 | 0.50 |
ENST00000400100.1
ENST00000400098.1 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr10_+_122610687 | 0.45 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr18_+_32820990 | 0.45 |
ENST00000601719.1
ENST00000591206.1 ENST00000330501.7 ENST00000261333.6 ENST00000355632.4 ENST00000585800.1 |
ZNF397
|
zinc finger protein 397 |
| chr6_+_144164455 | 0.44 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr15_+_75639372 | 0.43 |
ENST00000566313.1
ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr14_+_60712463 | 0.42 |
ENST00000325642.3
ENST00000529574.1 |
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr14_+_101361107 | 0.42 |
ENST00000553584.1
ENST00000554852.1 |
MEG8
|
maternally expressed 8 (non-protein coding) |
| chr6_-_3195981 | 0.41 |
ENST00000425384.2
ENST00000435043.2 |
RP1-40E16.9
|
RP1-40E16.9 |
| chr4_-_39034542 | 0.38 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr12_-_57328187 | 0.37 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr16_-_21442874 | 0.37 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr8_-_82359662 | 0.36 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr16_-_28303360 | 0.34 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr11_+_7618413 | 0.33 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr11_+_3011093 | 0.31 |
ENST00000332881.2
|
AC131971.1
|
HCG1782999; PRO0943; Uncharacterized protein |
| chr16_+_28303804 | 0.31 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr1_+_156698743 | 0.30 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr17_-_66951474 | 0.30 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr15_+_66585950 | 0.30 |
ENST00000525109.1
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr19_-_2783363 | 0.29 |
ENST00000221566.2
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr10_-_17243579 | 0.28 |
ENST00000525762.1
ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr5_+_175511859 | 0.28 |
ENST00000503724.2
ENST00000253490.4 |
FAM153B
|
family with sequence similarity 153, member B |
| chr16_+_22518495 | 0.27 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr1_+_100598691 | 0.27 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr10_-_71169031 | 0.27 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr19_-_21950332 | 0.26 |
ENST00000598026.1
|
ZNF100
|
zinc finger protein 100 |
| chr10_-_12084770 | 0.25 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr10_-_5652705 | 0.25 |
ENST00000425246.1
|
RP11-336A10.5
|
RP11-336A10.5 |
| chr4_-_4544061 | 0.25 |
ENST00000507908.1
|
STX18
|
syntaxin 18 |
| chr7_-_33148975 | 0.22 |
ENST00000297157.3
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr6_+_80129989 | 0.21 |
ENST00000429444.1
|
RP1-232L24.3
|
RP1-232L24.3 |
| chr1_+_156698708 | 0.18 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr2_+_87144738 | 0.18 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr19_-_2783255 | 0.18 |
ENST00000589251.1
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr17_-_39623681 | 0.16 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr12_+_64173583 | 0.15 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr12_+_55886147 | 0.15 |
ENST00000379662.1
ENST00000548615.1 |
OR6C68
|
olfactory receptor, family 6, subfamily C, member 68 |
| chr1_+_100598742 | 0.15 |
ENST00000370139.1
|
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr15_+_63414760 | 0.14 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr17_-_56082455 | 0.13 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr16_-_279405 | 0.10 |
ENST00000430864.1
ENST00000293872.8 ENST00000337351.4 ENST00000397783.1 |
LUC7L
|
LUC7-like (S. cerevisiae) |
| chr13_-_28024681 | 0.09 |
ENST00000381116.1
ENST00000381120.3 ENST00000431572.2 |
MTIF3
|
mitochondrial translational initiation factor 3 |
| chr12_-_123728548 | 0.09 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr10_-_96829246 | 0.08 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr12_-_39734783 | 0.08 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr4_+_69313145 | 0.07 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr8_-_19459993 | 0.06 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr3_-_122694215 | 0.05 |
ENST00000449546.1
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr7_-_143966381 | 0.04 |
ENST00000487179.1
|
CTAGE8
|
CTAGE family, member 8 |
| chr14_-_31856397 | 0.04 |
ENST00000538864.2
ENST00000550366.1 |
HEATR5A
|
HEAT repeat containing 5A |
| chr2_-_152118276 | 0.02 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.5 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.9 | 4.7 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.3 | 1.6 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.3 | 2.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.2 | 3.4 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.2 | 1.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 2.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 2.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 1.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.9 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 2.0 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.3 | GO:0035106 | negative regulation of luteinizing hormone secretion(GO:0033685) operant conditioning(GO:0035106) |
| 0.1 | 0.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 1.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 1.1 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0036024 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 14.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 4.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 1.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.5 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 13.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.5 | 3.4 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.3 | 2.1 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.3 | 1.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 1.2 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.2 | 0.7 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.2 | 4.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 1.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 2.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 1.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 1.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.9 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.3 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 2.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 2.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 2.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 7.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |