Project
Illumina Body Map 2
Navigation
Downloads
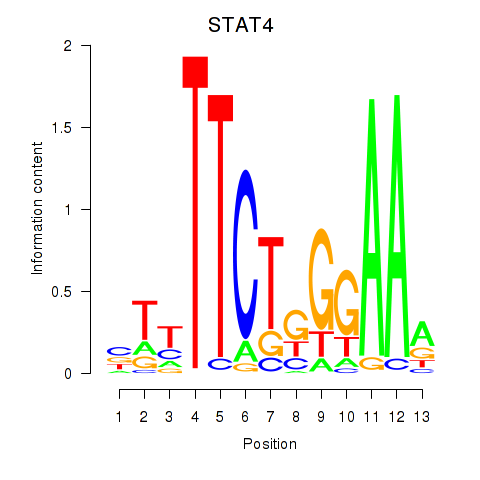
Results for STAT4
Z-value: 0.73
Transcription factors associated with STAT4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT4
|
ENSG00000138378.13 | signal transducer and activator of transcription 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT4 | hg19_v2_chr2_-_192015697_192015743 | -0.04 | 8.1e-01 | Click! |
Activity profile of STAT4 motif
Sorted Z-values of STAT4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_196242233 | 2.35 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr3_+_50649302 | 1.80 |
ENST00000446044.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr4_-_76957214 | 1.60 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr4_+_169575875 | 1.49 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_-_15082050 | 1.25 |
ENST00000540097.1
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr6_-_49712123 | 1.13 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr6_-_123958141 | 1.01 |
ENST00000334268.4
|
TRDN
|
triadin |
| chr6_-_49712072 | 1.00 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr16_-_46782221 | 0.98 |
ENST00000394809.4
|
MYLK3
|
myosin light chain kinase 3 |
| chr1_+_202431859 | 0.96 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr6_-_123958051 | 0.92 |
ENST00000546248.1
|
TRDN
|
triadin |
| chr6_-_49712091 | 0.91 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr10_+_115312766 | 0.87 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr3_-_108672742 | 0.77 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr6_-_123957942 | 0.77 |
ENST00000398178.3
|
TRDN
|
triadin |
| chr6_-_123958111 | 0.75 |
ENST00000542443.1
|
TRDN
|
triadin |
| chr4_-_177162822 | 0.75 |
ENST00000512254.1
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr3_-_108672609 | 0.73 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr9_-_134151915 | 0.68 |
ENST00000372271.3
|
FAM78A
|
family with sequence similarity 78, member A |
| chr1_-_120935894 | 0.66 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr1_+_149754227 | 0.64 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chrX_+_107288280 | 0.63 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_-_144994909 | 0.62 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr19_-_51875894 | 0.60 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chrX_+_107288239 | 0.60 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrX_+_107288197 | 0.60 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr12_-_11139511 | 0.59 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr1_+_161551101 | 0.58 |
ENST00000367962.4
ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr15_+_63340775 | 0.57 |
ENST00000559281.1
ENST00000317516.7 |
TPM1
|
tropomyosin 1 (alpha) |
| chr19_+_35225060 | 0.57 |
ENST00000599244.1
ENST00000392232.3 |
ZNF181
|
zinc finger protein 181 |
| chrX_-_46187069 | 0.56 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr14_+_22538811 | 0.52 |
ENST00000390450.3
|
TRAV22
|
T cell receptor alpha variable 22 |
| chr1_+_161632937 | 0.51 |
ENST00000236937.9
ENST00000367961.4 ENST00000358671.5 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr16_-_67517716 | 0.50 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr5_-_127674883 | 0.50 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr5_-_111092873 | 0.50 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr4_-_48116540 | 0.50 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr15_+_63340858 | 0.46 |
ENST00000560615.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr14_-_25103472 | 0.45 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr3_-_156534754 | 0.44 |
ENST00000472943.1
ENST00000473352.1 |
LINC00886
|
long intergenic non-protein coding RNA 886 |
| chr2_+_113885138 | 0.44 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr12_+_60083118 | 0.44 |
ENST00000261187.4
ENST00000543448.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_-_144994840 | 0.44 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_-_109655377 | 0.44 |
ENST00000369948.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr8_-_42358742 | 0.43 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr5_-_111093081 | 0.43 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr1_-_109655355 | 0.43 |
ENST00000369945.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr3_-_183735651 | 0.43 |
ENST00000427120.2
ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr14_-_100841930 | 0.43 |
ENST00000555031.1
ENST00000553395.1 ENST00000553545.1 ENST00000344102.5 ENST00000556338.1 ENST00000392882.2 ENST00000553934.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr14_-_100841670 | 0.43 |
ENST00000557297.1
ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr17_-_42994283 | 0.41 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr1_-_161337662 | 0.40 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr5_-_111093340 | 0.40 |
ENST00000508870.1
|
NREP
|
neuronal regeneration related protein |
| chr19_-_44405941 | 0.39 |
ENST00000587128.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr19_-_44405623 | 0.39 |
ENST00000591815.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr5_-_111092930 | 0.39 |
ENST00000257435.7
|
NREP
|
neuronal regeneration related protein |
| chr11_-_119999611 | 0.38 |
ENST00000529044.1
|
TRIM29
|
tripartite motif containing 29 |
| chrX_-_70474910 | 0.38 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr5_-_111093167 | 0.38 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr18_+_61144160 | 0.38 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr5_-_111093406 | 0.37 |
ENST00000379671.3
|
NREP
|
neuronal regeneration related protein |
| chr9_-_85882145 | 0.37 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr2_-_197675000 | 0.37 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr10_-_116444371 | 0.36 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr10_+_6625605 | 0.36 |
ENST00000414894.1
ENST00000449648.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr2_-_29297127 | 0.36 |
ENST00000331664.5
|
C2orf71
|
chromosome 2 open reading frame 71 |
| chr15_+_63340734 | 0.35 |
ENST00000560959.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr3_+_111260980 | 0.34 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr14_+_56127960 | 0.34 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr5_-_39274617 | 0.33 |
ENST00000510188.1
|
FYB
|
FYN binding protein |
| chr11_+_12302492 | 0.33 |
ENST00000533534.1
|
MICALCL
|
MICAL C-terminal like |
| chr9_-_116159635 | 0.32 |
ENST00000452726.1
|
ALAD
|
aminolevulinate dehydratase |
| chr12_+_14518598 | 0.31 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr16_+_78056412 | 0.31 |
ENST00000299642.4
ENST00000575655.1 |
CLEC3A
|
C-type lectin domain family 3, member A |
| chr6_+_32407619 | 0.30 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr5_-_64777733 | 0.30 |
ENST00000381055.3
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr5_+_36606700 | 0.30 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr6_-_56716686 | 0.30 |
ENST00000520645.1
|
DST
|
dystonin |
| chr11_+_5509915 | 0.30 |
ENST00000322641.5
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1 |
| chr11_-_119999539 | 0.29 |
ENST00000541857.1
|
TRIM29
|
tripartite motif containing 29 |
| chr18_-_67629015 | 0.29 |
ENST00000579496.1
|
CD226
|
CD226 molecule |
| chr2_-_160472952 | 0.29 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr2_+_234294585 | 0.29 |
ENST00000447484.1
|
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr14_-_100841794 | 0.28 |
ENST00000556295.1
ENST00000554820.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr6_+_84222220 | 0.28 |
ENST00000369700.3
|
PRSS35
|
protease, serine, 35 |
| chr1_+_117297007 | 0.28 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr14_+_56127989 | 0.28 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr7_-_115670804 | 0.27 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chrX_+_149861836 | 0.27 |
ENST00000542156.1
ENST00000370390.3 ENST00000490316.2 ENST00000445323.2 ENST00000544228.1 ENST00000451863.2 |
MTMR1
|
myotubularin related protein 1 |
| chr19_+_35225121 | 0.27 |
ENST00000595708.1
ENST00000593781.1 |
ZNF181
|
zinc finger protein 181 |
| chr20_+_5731027 | 0.26 |
ENST00000378979.4
ENST00000303142.6 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr6_+_134210243 | 0.26 |
ENST00000367882.4
|
TCF21
|
transcription factor 21 |
| chr12_-_11184006 | 0.26 |
ENST00000390675.2
|
TAS2R31
|
taste receptor, type 2, member 31 |
| chr1_+_17906970 | 0.26 |
ENST00000375415.1
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr17_-_65235916 | 0.26 |
ENST00000579861.1
|
HELZ
|
helicase with zinc finger |
| chr3_+_111260954 | 0.26 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr18_+_6729725 | 0.25 |
ENST00000400091.2
ENST00000583410.1 ENST00000584387.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr6_+_31082603 | 0.25 |
ENST00000259881.9
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr7_-_115670792 | 0.25 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr11_+_118175596 | 0.25 |
ENST00000528600.1
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr2_+_68962014 | 0.25 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr12_-_91398796 | 0.25 |
ENST00000261172.3
ENST00000551767.1 |
EPYC
|
epiphycan |
| chr5_-_140013224 | 0.25 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr9_-_95166841 | 0.25 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr9_-_95166884 | 0.24 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr5_-_64777685 | 0.24 |
ENST00000536360.1
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr11_-_133715394 | 0.24 |
ENST00000299140.3
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chr1_-_185597619 | 0.23 |
ENST00000608417.1
ENST00000436955.1 |
GS1-204I12.1
|
GS1-204I12.1 |
| chr3_-_183735731 | 0.23 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr6_+_32811861 | 0.23 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr1_-_8000872 | 0.22 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr2_+_27760247 | 0.22 |
ENST00000447166.1
|
AC109829.1
|
Uncharacterized protein |
| chr22_+_44761431 | 0.22 |
ENST00000406912.1
|
RP1-32I10.10
|
Uncharacterized protein |
| chr4_-_155533787 | 0.22 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr16_+_19467772 | 0.22 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr3_+_111260856 | 0.22 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr3_-_126327398 | 0.22 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr5_-_140013275 | 0.21 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr19_+_35168547 | 0.21 |
ENST00000502743.1
ENST00000509528.1 ENST00000506901.1 |
ZNF302
|
zinc finger protein 302 |
| chr19_-_44952635 | 0.21 |
ENST00000592308.1
ENST00000588931.1 ENST00000291187.4 |
ZNF229
|
zinc finger protein 229 |
| chr19_-_10613862 | 0.21 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr3_-_71114066 | 0.21 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr7_-_33842742 | 0.21 |
ENST00000420185.1
ENST00000440034.1 |
RP11-89N17.4
|
RP11-89N17.4 |
| chr12_+_113587558 | 0.21 |
ENST00000335621.6
|
CCDC42B
|
coiled-coil domain containing 42B |
| chr18_+_18822216 | 0.20 |
ENST00000269218.6
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr18_+_6729698 | 0.20 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr2_+_95831529 | 0.20 |
ENST00000295210.6
ENST00000453539.2 |
ZNF2
|
zinc finger protein 2 |
| chr19_-_56904799 | 0.20 |
ENST00000589895.1
ENST00000589143.1 ENST00000301310.4 ENST00000586929.1 |
ZNF582
|
zinc finger protein 582 |
| chr17_-_27038063 | 0.20 |
ENST00000439862.3
|
PROCA1
|
protein interacting with cyclin A1 |
| chr18_+_61143994 | 0.19 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr14_+_100240019 | 0.19 |
ENST00000556199.1
|
EML1
|
echinoderm microtubule associated protein like 1 |
| chr9_+_100069933 | 0.19 |
ENST00000529487.1
|
CCDC180
|
coiled-coil domain containing 180 |
| chr5_+_53686658 | 0.19 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr17_-_34207295 | 0.19 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr6_-_32821599 | 0.19 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr17_-_38210644 | 0.19 |
ENST00000394128.2
ENST00000394127.2 ENST00000356271.3 ENST00000535071.2 ENST00000580885.1 ENST00000543759.2 ENST00000537674.2 ENST00000580517.1 ENST00000578161.1 |
MED24
|
mediator complex subunit 24 |
| chr10_+_81065975 | 0.18 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr16_-_1660662 | 0.18 |
ENST00000569646.1
|
IFT140
|
intraflagellar transport 140 homolog (Chlamydomonas) |
| chr19_+_57106647 | 0.18 |
ENST00000328070.6
|
ZNF71
|
zinc finger protein 71 |
| chr14_+_55034599 | 0.17 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr2_-_84686552 | 0.16 |
ENST00000393868.2
|
SUCLG1
|
succinate-CoA ligase, alpha subunit |
| chr14_-_81408063 | 0.16 |
ENST00000557411.1
|
CEP128
|
centrosomal protein 128kDa |
| chr3_+_122296465 | 0.16 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr2_+_68961934 | 0.16 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr11_+_118175132 | 0.16 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr2_+_68961905 | 0.16 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr15_+_45021183 | 0.16 |
ENST00000559390.1
|
TRIM69
|
tripartite motif containing 69 |
| chr3_-_194119995 | 0.15 |
ENST00000323007.3
|
GP5
|
glycoprotein V (platelet) |
| chr5_-_95018660 | 0.15 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr6_-_116833500 | 0.15 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr19_+_57106624 | 0.15 |
ENST00000599599.1
|
ZNF71
|
zinc finger protein 71 |
| chr15_-_60690163 | 0.15 |
ENST00000558998.1
ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2
|
annexin A2 |
| chr6_+_139349903 | 0.15 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr14_+_20811722 | 0.15 |
ENST00000429687.3
|
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr10_+_6625733 | 0.15 |
ENST00000607982.1
ENST00000608526.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr1_-_226065330 | 0.15 |
ENST00000436966.1
|
TMEM63A
|
transmembrane protein 63A |
| chr17_-_58603482 | 0.14 |
ENST00000585368.1
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr5_+_137203541 | 0.14 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr12_-_55367331 | 0.14 |
ENST00000526532.1
ENST00000532757.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr10_-_6622201 | 0.14 |
ENST00000539722.1
ENST00000397176.2 |
PRKCQ
|
protein kinase C, theta |
| chr6_+_108977520 | 0.14 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr12_+_20848282 | 0.14 |
ENST00000545604.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr15_-_75660919 | 0.14 |
ENST00000569482.1
ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1
|
mannosidase, alpha, class 2C, member 1 |
| chr14_+_96722539 | 0.14 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr4_+_130017268 | 0.13 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr6_-_166581333 | 0.13 |
ENST00000366876.2
|
T
|
T, brachyury homolog (mouse) |
| chr19_+_35168633 | 0.13 |
ENST00000505365.2
|
ZNF302
|
zinc finger protein 302 |
| chr14_+_20811766 | 0.13 |
ENST00000250416.5
ENST00000527915.1 |
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr7_-_41740181 | 0.13 |
ENST00000442711.1
|
INHBA
|
inhibin, beta A |
| chr6_+_31540056 | 0.13 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr19_+_44669280 | 0.13 |
ENST00000590089.1
ENST00000454662.2 |
ZNF226
|
zinc finger protein 226 |
| chr21_+_38593701 | 0.12 |
ENST00000440629.1
|
AP001432.14
|
AP001432.14 |
| chr16_+_58283814 | 0.12 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr7_+_10000640 | 0.12 |
ENST00000451755.1
|
AC006373.1
|
AC006373.1 |
| chr1_-_144364246 | 0.12 |
ENST00000540273.1
|
PPIAL4B
|
peptidylprolyl isomerase A (cyclophilin A)-like 4B |
| chr9_-_95166976 | 0.12 |
ENST00000447356.1
|
OGN
|
osteoglycin |
| chr5_+_137203557 | 0.12 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr14_+_35591020 | 0.12 |
ENST00000603611.1
|
KIAA0391
|
KIAA0391 |
| chr5_+_137203465 | 0.12 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chrX_-_63005405 | 0.11 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr12_+_27396901 | 0.11 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr13_-_41240717 | 0.11 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr15_-_60690932 | 0.11 |
ENST00000559818.1
|
ANXA2
|
annexin A2 |
| chr14_-_35591156 | 0.11 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr9_-_116163400 | 0.11 |
ENST00000277315.5
ENST00000448137.1 ENST00000409155.3 |
ALAD
|
aminolevulinate dehydratase |
| chr1_+_149553003 | 0.11 |
ENST00000369222.3
|
PPIAL4C
|
peptidylprolyl isomerase A (cyclophilin A)-like 4C |
| chrX_+_54834159 | 0.11 |
ENST00000375053.2
ENST00000347546.4 ENST00000375062.4 |
MAGED2
|
melanoma antigen family D, 2 |
| chr19_+_50529212 | 0.11 |
ENST00000270617.3
ENST00000445728.3 ENST00000601364.1 |
ZNF473
|
zinc finger protein 473 |
| chr1_+_214163033 | 0.11 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr17_+_7358889 | 0.10 |
ENST00000575379.1
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chrX_+_24711997 | 0.10 |
ENST00000379068.3
ENST00000379059.3 |
POLA1
|
polymerase (DNA directed), alpha 1, catalytic subunit |
| chr5_-_16509101 | 0.10 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr10_-_50396357 | 0.10 |
ENST00000453436.1
ENST00000474718.1 |
C10orf128
|
chromosome 10 open reading frame 128 |
| chr20_+_5731083 | 0.10 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr3_+_130064359 | 0.10 |
ENST00000432398.2
ENST00000265379.6 |
COL6A5
|
collagen, type VI, alpha 5 |
| chr12_+_56435637 | 0.10 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr11_+_73003824 | 0.09 |
ENST00000538328.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr3_+_38179969 | 0.09 |
ENST00000396334.3
ENST00000417037.2 ENST00000424893.1 ENST00000495303.1 ENST00000443433.2 ENST00000421516.1 |
MYD88
|
myeloid differentiation primary response 88 |
| chr1_+_179699318 | 0.09 |
ENST00000423879.1
|
RP11-12M5.1
|
RP11-12M5.1 |
| chr19_+_35168567 | 0.09 |
ENST00000457781.2
ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302
|
zinc finger protein 302 |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 0.4 | 1.1 | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.3 | 0.8 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.2 | 1.0 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 1.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 1.6 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 1.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.4 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 1.5 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.3 | GO:0060435 | branchiomeric skeletal muscle development(GO:0014707) bronchiole development(GO:0060435) |
| 0.1 | 0.4 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.5 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.5 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 0.3 | GO:0044115 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.1 | 1.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.5 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 1.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.3 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.2 | GO:2000110 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.3 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.2 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.1 | GO:0061114 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.7 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.3 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0070417 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) cellular response to cold(GO:0070417) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:1902230 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902229) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.3 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.3 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 1.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 3.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.6 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 1.0 | GO:0031672 | A band(GO:0031672) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 1.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 1.5 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 1.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 0.7 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.4 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.4 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 1.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.5 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.2 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |