Project
Illumina Body Map 2
Navigation
Downloads
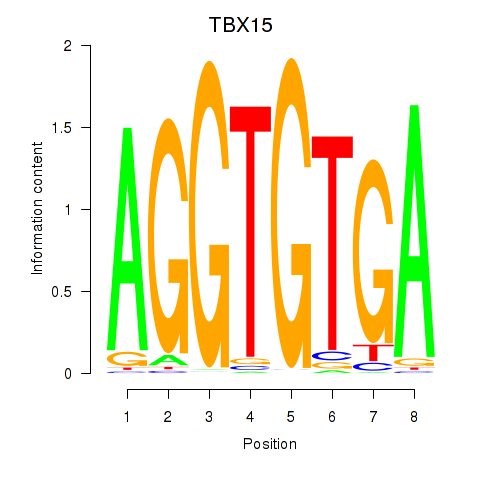
Results for TBX15_MGA
Z-value: 1.52
Transcription factors associated with TBX15_MGA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX15
|
ENSG00000092607.9 | T-box transcription factor 15 |
|
MGA
|
ENSG00000174197.12 | MAX dimerization protein MGA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX15 | hg19_v2_chr1_-_119530428_119530572 | 0.10 | 5.7e-01 | Click! |
| MGA | hg19_v2_chr15_+_41952591_41952672 | 0.03 | 8.6e-01 | Click! |
Activity profile of TBX15_MGA motif
Sorted Z-values of TBX15_MGA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_128788914 | 5.26 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr1_-_37499726 | 5.01 |
ENST00000373091.3
ENST00000373093.4 |
GRIK3
|
glutamate receptor, ionotropic, kainate 3 |
| chr20_+_44098385 | 4.91 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr1_-_85930246 | 4.86 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr20_+_44098346 | 4.81 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr4_-_186732048 | 4.48 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_+_95078714 | 4.48 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr9_-_34397800 | 4.24 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr1_+_38022572 | 4.20 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr1_+_38022513 | 4.15 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr3_+_120626919 | 4.15 |
ENST00000273666.6
ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L
|
syntaxin binding protein 5-like |
| chr1_-_44482979 | 3.97 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr15_+_90319557 | 3.89 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chr9_+_140032842 | 3.76 |
ENST00000371561.3
ENST00000315048.3 |
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| chr9_+_139847347 | 3.72 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chrX_+_70443050 | 3.64 |
ENST00000361726.6
|
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr19_-_7990991 | 3.55 |
ENST00000318978.4
|
CTXN1
|
cortexin 1 |
| chr2_-_61108449 | 3.55 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chrX_+_122318318 | 3.51 |
ENST00000371251.1
ENST00000371256.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr19_+_46531127 | 3.51 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chr16_-_81110683 | 3.40 |
ENST00000565253.1
ENST00000378611.4 ENST00000299578.5 |
C16orf46
|
chromosome 16 open reading frame 46 |
| chr7_+_97361388 | 3.39 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chr11_+_73358690 | 3.37 |
ENST00000545798.1
ENST00000539157.1 ENST00000546251.1 ENST00000535582.1 ENST00000538227.1 ENST00000543524.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr3_+_167453493 | 3.35 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr4_-_186456652 | 3.34 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr12_+_56473939 | 3.33 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chrX_+_122318006 | 3.33 |
ENST00000371266.1
ENST00000264357.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr9_+_71986182 | 3.30 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr18_+_31185530 | 3.29 |
ENST00000586327.1
|
ASXL3
|
additional sex combs like 3 (Drosophila) |
| chrX_+_122318224 | 3.27 |
ENST00000542149.1
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr7_-_81635106 | 3.26 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr5_-_132112907 | 3.25 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr1_-_153066998 | 3.25 |
ENST00000368750.3
|
SPRR2E
|
small proline-rich protein 2E |
| chr3_-_93747425 | 3.24 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr5_-_132112921 | 3.20 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr11_+_73358594 | 3.09 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr4_-_186456766 | 3.08 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr20_+_34680620 | 3.05 |
ENST00000430276.1
ENST00000373950.2 ENST00000452261.1 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr4_-_44450814 | 3.04 |
ENST00000360029.3
|
KCTD8
|
potassium channel tetramerization domain containing 8 |
| chr11_-_84028180 | 3.03 |
ENST00000280241.8
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chrX_+_122318113 | 3.02 |
ENST00000371264.3
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr1_-_226129083 | 3.01 |
ENST00000420304.2
|
LEFTY2
|
left-right determination factor 2 |
| chr5_+_113697983 | 3.01 |
ENST00000264773.3
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr18_-_30050395 | 2.98 |
ENST00000269209.6
ENST00000399218.4 |
GAREM
|
GRB2 associated, regulator of MAPK1 |
| chr12_-_120805872 | 2.95 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr1_-_226129189 | 2.95 |
ENST00000366820.5
|
LEFTY2
|
left-right determination factor 2 |
| chr14_+_85994943 | 2.89 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr5_+_140571902 | 2.88 |
ENST00000239446.4
|
PCDHB10
|
protocadherin beta 10 |
| chr14_+_78870030 | 2.87 |
ENST00000553631.1
ENST00000554719.1 |
NRXN3
|
neurexin 3 |
| chr3_+_167453026 | 2.85 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr17_+_37782955 | 2.85 |
ENST00000580825.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chrX_+_102862834 | 2.83 |
ENST00000372627.5
ENST00000243286.3 |
TCEAL3
|
transcription elongation factor A (SII)-like 3 |
| chr2_-_27486951 | 2.78 |
ENST00000432351.1
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr8_+_75736761 | 2.76 |
ENST00000260113.2
|
PI15
|
peptidase inhibitor 15 |
| chr7_-_123673471 | 2.72 |
ENST00000455783.1
|
TMEM229A
|
transmembrane protein 229A |
| chr17_+_77021702 | 2.70 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr9_+_102584128 | 2.65 |
ENST00000338488.4
ENST00000395097.2 |
NR4A3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr11_+_46402297 | 2.62 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr2_-_217560248 | 2.62 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr5_+_125800836 | 2.60 |
ENST00000511134.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr7_+_97361218 | 2.59 |
ENST00000319273.5
|
TAC1
|
tachykinin, precursor 1 |
| chr1_-_111174054 | 2.58 |
ENST00000369770.3
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr10_+_45406627 | 2.55 |
ENST00000389583.4
|
TMEM72
|
transmembrane protein 72 |
| chr3_-_120170052 | 2.54 |
ENST00000295633.3
|
FSTL1
|
follistatin-like 1 |
| chrX_+_101380642 | 2.53 |
ENST00000372780.1
ENST00000329035.2 |
TCEAL2
|
transcription elongation factor A (SII)-like 2 |
| chrX_-_55515635 | 2.53 |
ENST00000500968.3
|
USP51
|
ubiquitin specific peptidase 51 |
| chr2_+_62933001 | 2.53 |
ENST00000263991.5
ENST00000354487.3 |
EHBP1
|
EH domain binding protein 1 |
| chr15_-_83315874 | 2.50 |
ENST00000569257.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr3_-_100712352 | 2.47 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr1_-_11907829 | 2.43 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr1_-_230561475 | 2.42 |
ENST00000391860.1
|
PGBD5
|
piggyBac transposable element derived 5 |
| chr3_-_48471454 | 2.36 |
ENST00000296440.6
ENST00000448774.2 |
PLXNB1
|
plexin B1 |
| chr15_+_76030311 | 2.29 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr6_+_121756809 | 2.25 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr11_+_117947782 | 2.23 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr1_+_8378140 | 2.23 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr2_+_202937972 | 2.18 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr17_-_10707410 | 2.15 |
ENST00000581851.1
|
LINC00675
|
long intergenic non-protein coding RNA 675 |
| chr14_+_51026926 | 2.15 |
ENST00000557735.1
|
ATL1
|
atlastin GTPase 1 |
| chr14_+_51026844 | 2.14 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr11_-_83393429 | 2.14 |
ENST00000426717.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr4_-_66535653 | 2.12 |
ENST00000354839.4
ENST00000432638.2 |
EPHA5
|
EPH receptor A5 |
| chr1_+_33219592 | 2.11 |
ENST00000373481.3
|
KIAA1522
|
KIAA1522 |
| chr2_-_27718052 | 2.09 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr1_+_110693103 | 2.07 |
ENST00000331565.4
|
SLC6A17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr7_-_120497178 | 2.07 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr2_+_62932779 | 2.05 |
ENST00000427809.1
ENST00000405482.1 ENST00000431489.1 |
EHBP1
|
EH domain binding protein 1 |
| chr15_-_90294523 | 2.05 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr18_-_59561417 | 2.02 |
ENST00000591306.1
|
RNF152
|
ring finger protein 152 |
| chr17_-_74533734 | 2.02 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr2_-_175869936 | 2.00 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chr20_+_43343476 | 1.96 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr6_+_108616243 | 1.92 |
ENST00000421954.1
|
LACE1
|
lactation elevated 1 |
| chr20_+_2517253 | 1.88 |
ENST00000358864.1
|
TMC2
|
transmembrane channel-like 2 |
| chr7_-_150675372 | 1.88 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr5_-_124080203 | 1.87 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr13_-_36429763 | 1.86 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr10_+_119184702 | 1.86 |
ENST00000549104.1
|
CTA-109P11.4
|
CTA-109P11.4 |
| chr20_+_43343886 | 1.84 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr3_-_100712292 | 1.83 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr4_+_57371509 | 1.82 |
ENST00000360096.2
|
ARL9
|
ADP-ribosylation factor-like 9 |
| chr4_-_66536057 | 1.80 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr4_-_146859787 | 1.79 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr7_+_107110488 | 1.78 |
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22 |
| chr11_-_125932685 | 1.78 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr14_+_33408449 | 1.78 |
ENST00000346562.2
ENST00000341321.4 ENST00000548645.1 ENST00000356141.4 ENST00000357798.5 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr15_+_74466744 | 1.76 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr4_-_73434498 | 1.76 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr10_-_104179682 | 1.72 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr1_+_160051319 | 1.71 |
ENST00000368088.3
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr19_-_46974741 | 1.71 |
ENST00000313683.10
ENST00000602246.1 |
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr20_-_22565101 | 1.70 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chr15_-_83316254 | 1.69 |
ENST00000567678.1
ENST00000450751.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr16_+_3162557 | 1.68 |
ENST00000382192.3
ENST00000219091.4 ENST00000444510.2 ENST00000414351.1 |
ZNF205
|
zinc finger protein 205 |
| chr15_-_83316087 | 1.68 |
ENST00000568757.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr17_-_1619535 | 1.68 |
ENST00000573075.1
ENST00000574306.1 |
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr6_+_32132360 | 1.67 |
ENST00000333845.6
ENST00000395512.1 ENST00000432129.1 |
EGFL8
|
EGF-like-domain, multiple 8 |
| chr18_+_32073839 | 1.67 |
ENST00000590412.1
|
DTNA
|
dystrobrevin, alpha |
| chr2_+_43864387 | 1.66 |
ENST00000282406.4
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr2_-_175870085 | 1.64 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr3_-_49459878 | 1.64 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
| chr6_-_86099898 | 1.63 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chrX_-_10851762 | 1.63 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr11_-_6502580 | 1.62 |
ENST00000423813.2
ENST00000396777.3 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr8_-_99955042 | 1.61 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr19_-_46974664 | 1.61 |
ENST00000438932.2
|
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr11_+_66790816 | 1.60 |
ENST00000527043.1
|
SYT12
|
synaptotagmin XII |
| chr12_-_57634475 | 1.60 |
ENST00000393825.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr3_+_4535155 | 1.60 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr11_+_117947724 | 1.60 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr5_+_94727048 | 1.59 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr3_-_66551397 | 1.58 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr2_+_166150541 | 1.58 |
ENST00000283256.6
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chrX_-_138287168 | 1.57 |
ENST00000436198.1
|
FGF13
|
fibroblast growth factor 13 |
| chr20_-_55841398 | 1.56 |
ENST00000395864.3
|
BMP7
|
bone morphogenetic protein 7 |
| chr2_+_176972000 | 1.55 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr5_-_139283982 | 1.55 |
ENST00000340391.3
|
NRG2
|
neuregulin 2 |
| chr2_-_157189180 | 1.54 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chrX_-_99891796 | 1.54 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr19_+_54926621 | 1.54 |
ENST00000376530.3
ENST00000445095.1 ENST00000391739.3 ENST00000376531.3 |
TTYH1
|
tweety family member 1 |
| chr21_-_42880075 | 1.54 |
ENST00000332149.5
|
TMPRSS2
|
transmembrane protease, serine 2 |
| chr2_-_39664405 | 1.54 |
ENST00000341681.5
ENST00000263881.3 |
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr13_-_48575443 | 1.53 |
ENST00000378654.3
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr12_+_50451331 | 1.53 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr12_+_50451462 | 1.53 |
ENST00000447966.2
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr20_-_55841662 | 1.49 |
ENST00000395863.3
ENST00000450594.2 |
BMP7
|
bone morphogenetic protein 7 |
| chr18_+_32073253 | 1.47 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr11_+_124789146 | 1.46 |
ENST00000408930.5
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1 |
| chr13_-_48575401 | 1.46 |
ENST00000433022.1
ENST00000544100.1 |
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr3_-_49459865 | 1.45 |
ENST00000427987.1
|
AMT
|
aminomethyltransferase |
| chr4_-_168155700 | 1.44 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_+_142315225 | 1.44 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr13_-_48575376 | 1.44 |
ENST00000434484.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr16_-_28550320 | 1.43 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr4_+_111397216 | 1.43 |
ENST00000265162.5
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A) |
| chr14_-_45603657 | 1.43 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr11_-_119993979 | 1.42 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr14_+_51026743 | 1.42 |
ENST00000358385.6
ENST00000357032.3 ENST00000354525.4 |
ATL1
|
atlastin GTPase 1 |
| chr11_-_57004658 | 1.42 |
ENST00000606794.1
|
APLNR
|
apelin receptor |
| chr17_-_10741762 | 1.40 |
ENST00000580256.2
|
PIRT
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr4_-_66536196 | 1.39 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr1_-_111148241 | 1.39 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr5_-_9630463 | 1.39 |
ENST00000382492.2
|
TAS2R1
|
taste receptor, type 2, member 1 |
| chr15_+_74421961 | 1.38 |
ENST00000565159.1
ENST00000567206.1 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr12_+_27849378 | 1.37 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chrX_+_69642881 | 1.37 |
ENST00000453994.2
ENST00000536730.1 ENST00000538649.1 ENST00000374382.3 |
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr11_-_27494309 | 1.36 |
ENST00000389858.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr21_-_39870339 | 1.36 |
ENST00000429727.2
ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr6_+_39760129 | 1.36 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr21_-_42879909 | 1.36 |
ENST00000458356.1
ENST00000398585.3 ENST00000424093.1 |
TMPRSS2
|
transmembrane protease, serine 2 |
| chr2_+_210636697 | 1.36 |
ENST00000439458.1
ENST00000272845.6 |
UNC80
|
unc-80 homolog (C. elegans) |
| chr11_+_6260298 | 1.36 |
ENST00000379936.2
|
CNGA4
|
cyclic nucleotide gated channel alpha 4 |
| chr7_-_44179972 | 1.36 |
ENST00000446581.1
|
MYL7
|
myosin, light chain 7, regulatory |
| chrX_-_133119476 | 1.35 |
ENST00000543339.1
|
GPC3
|
glypican 3 |
| chr6_+_108616054 | 1.33 |
ENST00000368977.4
|
LACE1
|
lactation elevated 1 |
| chr17_-_1619491 | 1.32 |
ENST00000570416.1
ENST00000575626.1 ENST00000610106.1 ENST00000608198.1 ENST00000609442.1 ENST00000334146.3 ENST00000576489.1 ENST00000608245.1 ENST00000609398.1 ENST00000608913.1 ENST00000574016.1 ENST00000571091.1 ENST00000573127.1 ENST00000609990.1 ENST00000576749.1 |
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr19_+_54926601 | 1.32 |
ENST00000301194.4
|
TTYH1
|
tweety family member 1 |
| chr11_-_6502534 | 1.31 |
ENST00000254584.2
ENST00000525235.1 ENST00000445086.2 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr19_-_36001286 | 1.31 |
ENST00000602679.1
ENST00000492341.2 ENST00000472252.2 ENST00000602781.1 ENST00000402589.2 ENST00000458071.1 ENST00000436012.1 ENST00000443640.1 ENST00000450261.1 ENST00000467637.1 ENST00000480502.1 ENST00000474928.1 ENST00000414866.2 ENST00000392206.2 ENST00000488892.1 |
DMKN
|
dermokine |
| chr4_-_168155577 | 1.30 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_-_66551351 | 1.30 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr5_-_172756506 | 1.29 |
ENST00000265087.4
|
STC2
|
stanniocalcin 2 |
| chr5_-_34043310 | 1.29 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr15_-_88799384 | 1.29 |
ENST00000540489.2
ENST00000557856.1 ENST00000558676.1 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr8_+_102504651 | 1.28 |
ENST00000251808.3
ENST00000521085.1 |
GRHL2
|
grainyhead-like 2 (Drosophila) |
| chr17_+_39975544 | 1.28 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chrX_+_105937068 | 1.27 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr5_+_68788594 | 1.27 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr5_-_138780159 | 1.27 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr9_-_73483958 | 1.26 |
ENST00000377101.1
ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr20_+_43343517 | 1.26 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr1_+_35734616 | 1.26 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4 |
| chr19_+_35773242 | 1.25 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr9_+_133986782 | 1.25 |
ENST00000372301.2
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr18_+_73118967 | 1.25 |
ENST00000578340.1
|
RP11-321M21.3
|
HCG1996301; Uncharacterized protein |
| chr3_+_142315294 | 1.24 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr1_-_182641037 | 1.23 |
ENST00000483095.2
|
RGS8
|
regulator of G-protein signaling 8 |
| chr3_+_63805017 | 1.23 |
ENST00000295896.8
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr2_-_27498186 | 1.23 |
ENST00000447008.2
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr12_-_48152428 | 1.23 |
ENST00000449771.2
ENST00000395358.3 |
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX15_MGA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 1.8 | 5.3 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 1.3 | 4.0 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.3 | 3.8 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 1.1 | 4.4 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.9 | 2.6 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.9 | 2.6 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.8 | 2.5 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.8 | 3.0 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) allantois development(GO:1905069) |
| 0.7 | 2.1 | GO:0090381 | regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 | 4.0 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.6 | 1.9 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.6 | 2.5 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.6 | 3.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.6 | 2.4 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.6 | 3.6 | GO:0030421 | defecation(GO:0030421) |
| 0.6 | 0.6 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.6 | 2.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.6 | 2.3 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.6 | 2.8 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.5 | 1.6 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.5 | 2.4 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.5 | 1.5 | GO:0003168 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.4 | 3.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.4 | 1.7 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 | 1.3 | GO:0043132 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.4 | 1.2 | GO:1990641 | cellular response to bile acid(GO:1903413) response to iron ion starvation(GO:1990641) |
| 0.4 | 3.3 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.4 | 2.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.4 | 1.5 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.4 | 1.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.4 | 3.7 | GO:0015820 | leucine transport(GO:0015820) |
| 0.4 | 2.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 2.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.3 | 6.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 2.7 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 2.0 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.3 | 2.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.3 | 1.3 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.3 | 5.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.3 | 2.2 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.3 | 0.9 | GO:1901147 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.3 | 1.8 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 1.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.3 | 2.0 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.3 | 2.0 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 4.0 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.3 | 2.0 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.3 | 1.7 | GO:0010728 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.3 | 1.9 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.3 | 1.6 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.3 | 2.9 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.3 | 3.0 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.2 | 1.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 4.9 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 15.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 0.7 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 1.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 4.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 5.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 0.6 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 1.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 0.8 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.2 | 0.8 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.2 | 1.0 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 1.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 1.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.2 | 1.3 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.2 | 3.6 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 4.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.7 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 0.9 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 1.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 1.4 | GO:0044254 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.2 | 1.8 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 9.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.2 | 0.9 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.2 | 4.9 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.2 | 1.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 0.7 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.2 | 0.3 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.2 | 0.7 | GO:0061349 | chemoattraction of serotonergic neuron axon(GO:0036517) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.2 | 2.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 | 4.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 1.2 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.2 | 1.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 0.7 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.4 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.1 | 2.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.4 | GO:0070904 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 1.1 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 2.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 1.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 3.0 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.8 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.8 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 1.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 1.3 | GO:0071638 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 2.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.4 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.4 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.9 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.1 | 0.6 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 1.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 1.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 4.6 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.9 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 1.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.1 | GO:0021767 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.1 | 0.4 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 3.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.6 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 1.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.5 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.6 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 0.3 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.8 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 1.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 1.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 4.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 3.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.5 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.1 | 1.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.8 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.9 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.4 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.3 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.1 | 2.9 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 7.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.4 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.6 | GO:1903593 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of histamine secretion by mast cell(GO:1903593) |
| 0.1 | 0.2 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 2.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 4.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 2.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.3 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.3 | GO:0050893 | sensory processing(GO:0050893) |
| 0.1 | 0.9 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 0.7 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 4.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.4 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 0.4 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 1.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.3 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.2 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 3.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.3 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 0.5 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.8 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 2.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 2.6 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 1.5 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.1 | 0.5 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 1.0 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.8 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 2.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.1 | 0.6 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.1 | 0.4 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 1.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.8 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 2.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.1 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.0 | 0.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 1.4 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 0.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 2.7 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.4 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.7 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.5 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 1.0 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.9 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 1.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 2.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.9 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 3.6 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.9 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.4 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 1.1 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.5 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 3.1 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 2.2 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 1.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 14.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.7 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 2.0 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.8 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.6 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 1.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 1.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 1.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 2.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.4 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.9 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.4 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.6 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 1.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 1.5 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 3.2 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.6 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 1.0 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 1.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 1.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.5 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 1.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 5.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.4 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.4 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 2.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 5.0 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 0.5 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 1.2 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 5.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.4 | 5.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.3 | 2.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 13.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 2.7 | GO:0042025 | host cell nucleus(GO:0042025) |
| 0.3 | 14.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 2.9 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 0.8 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.2 | 2.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 6.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 0.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 3.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.9 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 2.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 5.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.5 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 2.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 3.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.6 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.9 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 8.0 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 3.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 3.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 3.6 | GO:0042995 | cell projection(GO:0042995) |
| 0.1 | 0.8 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 1.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 11.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 2.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.5 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 1.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.9 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 1.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 5.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 5.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 3.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 14.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 2.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 7.2 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 6.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 4.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 3.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 4.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 19.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.0 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 15.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 2.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.4 | GO:0030914 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.0 | 3.8 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 4.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 3.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 3.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 4.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 2.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.7 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 1.3 | 4.0 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 1.1 | 3.3 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.0 | 4.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.9 | 5.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.9 | 4.4 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.8 | 14.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.8 | 3.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.7 | 4.9 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.6 | 5.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.6 | 2.9 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.6 | 1.7 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.5 | 1.6 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.5 | 3.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.5 | 2.0 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.5 | 3.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.5 | 2.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.4 | 5.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.4 | 1.3 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.4 | 3.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.4 | 8.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 1.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.3 | 1.3 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.3 | 2.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 6.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 2.0 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.3 | 13.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.3 | 0.8 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.3 | 0.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 3.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 0.7 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 0.9 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 0.7 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.2 | 2.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 0.7 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 1.1 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.2 | 3.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 3.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 7.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 1.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 1.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 0.6 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 0.7 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.2 | 5.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 1.1 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.2 | 8.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 7.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 4.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 3.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 0.5 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.2 | 0.7 | GO:0005115 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 0.8 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 0.6 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 1.9 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 2.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 4.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.4 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 1.0 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 3.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.9 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.1 | 3.0 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 1.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.5 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 1.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 3.9 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.3 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.4 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.1 | 0.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 1.9 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.9 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.5 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 2.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.8 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.5 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.1 | 4.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 1.7 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 1.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.2 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 1.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 4.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.6 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 2.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.7 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 6.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 6.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 3.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 9.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 13.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.7 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 2.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 1.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 1.8 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) |
| 0.0 | 2.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 5.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.4 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 2.1 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 1.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 2.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 12.4 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 1.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 3.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 2.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 1.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 2.0 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 5.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 4.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 4.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 3.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 14.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 2.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 6.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 6.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.5 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 18.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 5.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 6.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 6.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 4.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 4.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 5.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 4.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 8.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 4.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 3.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 4.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 2.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 3.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 4.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 2.8 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.0 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 2.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 3.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.9 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 2.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |