Project
Illumina Body Map 2
Navigation
Downloads
Results for TBX20
Z-value: 0.98
Transcription factors associated with TBX20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX20
|
ENSG00000164532.10 | T-box transcription factor 20 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX20 | hg19_v2_chr7_-_35293740_35293758 | -0.23 | 2.0e-01 | Click! |
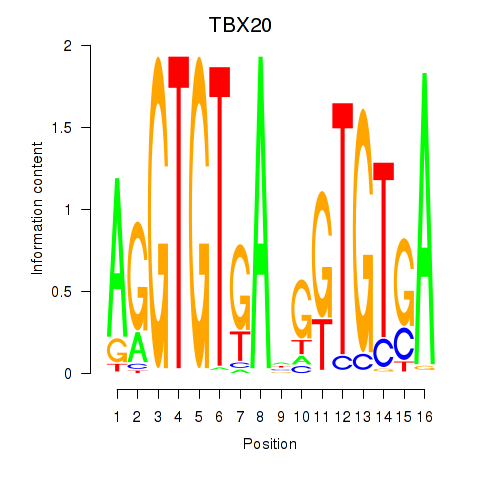
Activity profile of TBX20 motif
Sorted Z-values of TBX20 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_57662419 | 12.35 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_+_57662527 | 9.50 |
ENST00000563374.1
ENST00000568234.1 ENST00000565770.1 ENST00000564338.1 ENST00000566164.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_+_57662596 | 8.47 |
ENST00000567397.1
ENST00000568979.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr17_-_7080227 | 4.42 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr17_-_3599327 | 2.98 |
ENST00000551178.1
ENST00000552276.1 ENST00000547178.1 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr17_-_3599492 | 2.95 |
ENST00000435558.1
ENST00000345901.3 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr7_-_142099977 | 2.63 |
ENST00000390359.3
|
TRBV7-8
|
T cell receptor beta variable 7-8 |
| chr7_-_142207004 | 2.44 |
ENST00000426318.2
|
TRBV10-2
|
T cell receptor beta variable 10-2 |
| chr17_-_3599696 | 2.33 |
ENST00000225328.5
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr8_+_27183033 | 2.25 |
ENST00000420218.2
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr8_+_27182862 | 2.24 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chrX_+_78200913 | 2.22 |
ENST00000171757.2
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10 |
| chrX_+_78200829 | 2.02 |
ENST00000544091.1
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10 |
| chr12_+_15475331 | 1.80 |
ENST00000281171.4
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr12_+_15475462 | 1.72 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr15_-_22448819 | 1.72 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr13_-_30948036 | 1.46 |
ENST00000447147.1
ENST00000444319.1 |
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr5_+_133861339 | 1.31 |
ENST00000282605.4
ENST00000361895.2 ENST00000402835.1 |
PHF15
|
jade family PHD finger 2 |
| chr2_+_61108771 | 1.28 |
ENST00000394479.3
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr6_-_2842219 | 1.19 |
ENST00000380739.5
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr1_-_158300747 | 1.18 |
ENST00000451207.1
|
CD1B
|
CD1b molecule |
| chr19_-_4540486 | 1.16 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr6_-_2842087 | 1.16 |
ENST00000537185.1
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr14_+_77228532 | 1.14 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr2_+_61108650 | 1.02 |
ENST00000295025.8
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr7_-_139763521 | 0.94 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr12_-_121477039 | 0.83 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr12_-_121476959 | 0.81 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr5_+_135394840 | 0.80 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr4_+_160025300 | 0.79 |
ENST00000505478.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr3_-_183273477 | 0.76 |
ENST00000341319.3
|
KLHL6
|
kelch-like family member 6 |
| chr3_+_161214596 | 0.74 |
ENST00000327928.4
|
OTOL1
|
otolin 1 |
| chr3_+_186915274 | 0.72 |
ENST00000312295.4
|
RTP1
|
receptor (chemosensory) transporter protein 1 |
| chrX_+_67913471 | 0.69 |
ENST00000374597.3
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr5_+_161494521 | 0.68 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr19_+_7745708 | 0.67 |
ENST00000596148.1
ENST00000317378.5 ENST00000426877.2 |
TRAPPC5
|
trafficking protein particle complex 5 |
| chr6_-_90062543 | 0.64 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr1_+_87797351 | 0.63 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr12_-_121476750 | 0.55 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr17_-_76824975 | 0.51 |
ENST00000586066.2
|
USP36
|
ubiquitin specific peptidase 36 |
| chr22_-_46283597 | 0.51 |
ENST00000451118.1
|
WI2-85898F10.1
|
WI2-85898F10.1 |
| chr3_-_187694195 | 0.41 |
ENST00000446091.1
|
RP11-132N15.3
|
RP11-132N15.3 |
| chr7_+_30185496 | 0.37 |
ENST00000455738.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr6_-_107077347 | 0.36 |
ENST00000369063.3
ENST00000539449.1 |
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr21_-_45960078 | 0.35 |
ENST00000400375.1
|
KRTAP10-1
|
keratin associated protein 10-1 |
| chr19_+_35810164 | 0.32 |
ENST00000598537.1
|
CD22
|
CD22 molecule |
| chr19_-_10213335 | 0.30 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr14_-_106453155 | 0.30 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr20_+_44509857 | 0.30 |
ENST00000372523.1
ENST00000372520.1 |
ZSWIM1
|
zinc finger, SWIM-type containing 1 |
| chr15_-_38519066 | 0.29 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr11_-_17410629 | 0.29 |
ENST00000526912.1
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr21_-_46000481 | 0.28 |
ENST00000400372.1
|
KRTAP10-5
|
keratin associated protein 10-5 |
| chr4_-_26492076 | 0.26 |
ENST00000295589.3
|
CCKAR
|
cholecystokinin A receptor |
| chr1_+_147013182 | 0.25 |
ENST00000234739.3
|
BCL9
|
B-cell CLL/lymphoma 9 |
| chr19_-_9325542 | 0.24 |
ENST00000308682.2
|
OR7D4
|
olfactory receptor, family 7, subfamily D, member 4 |
| chr1_-_11118896 | 0.23 |
ENST00000465788.1
|
SRM
|
spermidine synthase |
| chr15_-_74421477 | 0.23 |
ENST00000514871.1
|
RP11-247C2.2
|
HCG2004779; Uncharacterized protein |
| chr20_-_48782639 | 0.21 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr10_+_88414338 | 0.14 |
ENST00000241891.5
ENST00000443292.1 |
OPN4
|
opsin 4 |
| chr22_+_38219389 | 0.14 |
ENST00000249041.2
|
GALR3
|
galanin receptor 3 |
| chrX_-_16881144 | 0.13 |
ENST00000416035.1
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr10_+_88414298 | 0.12 |
ENST00000372071.2
|
OPN4
|
opsin 4 |
| chr7_+_30185406 | 0.12 |
ENST00000324489.5
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr5_-_89825328 | 0.10 |
ENST00000500869.2
ENST00000315948.6 ENST00000509384.1 |
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr2_+_207630081 | 0.10 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr19_+_17579556 | 0.07 |
ENST00000442725.1
|
SLC27A1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr7_-_33140498 | 0.06 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr8_+_142264664 | 0.06 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr3_-_50396978 | 0.00 |
ENST00000266025.3
|
TMEM115
|
transmembrane protein 115 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX20
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.6 | 4.5 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.3 | 1.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 12.5 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 0.8 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 2.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.2 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.1 | 0.6 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 4.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.6 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 1.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.8 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 1.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 2.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.2 | 4.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 2.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 3.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.7 | 8.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.5 | 4.4 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.2 | 2.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 4.2 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.1 | 1.2 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 0.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 3.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 0.8 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 2.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 3.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 4.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 2.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |