Project
Illumina Body Map 2
Navigation
Downloads
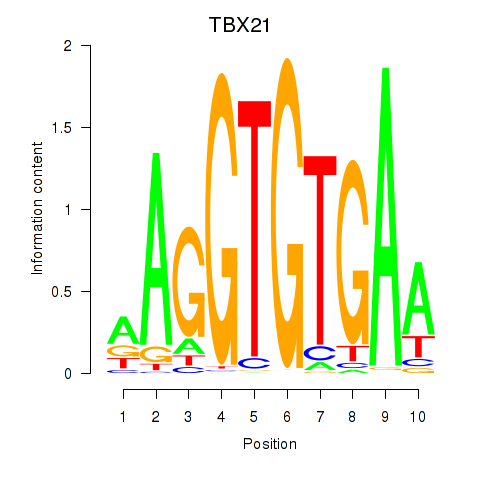
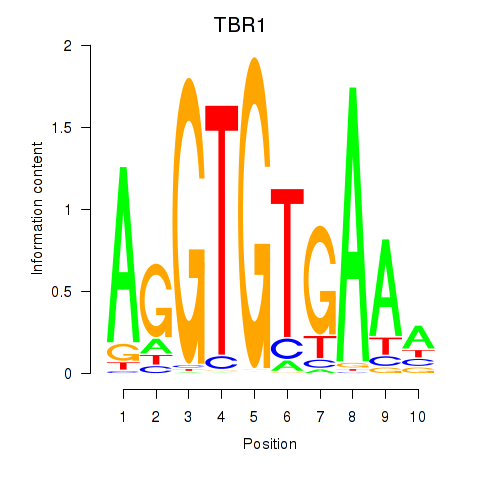
Results for TBX21_TBR1
Z-value: 1.09
Transcription factors associated with TBX21_TBR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX21
|
ENSG00000073861.2 | T-box transcription factor 21 |
|
TBR1
|
ENSG00000136535.10 | T-box brain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX21 | hg19_v2_chr17_+_45810594_45810610 | 0.38 | 3.0e-02 | Click! |
| TBR1 | hg19_v2_chr2_+_162272605_162272753 | -0.03 | 8.7e-01 | Click! |
Activity profile of TBX21_TBR1 motif
Sorted Z-values of TBX21_TBR1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_1993173 | 4.88 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr14_-_23876801 | 4.53 |
ENST00000356287.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr2_-_219031709 | 4.16 |
ENST00000295683.2
|
CXCR1
|
chemokine (C-X-C motif) receptor 1 |
| chr21_+_30503282 | 3.95 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr11_+_112832202 | 3.58 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr22_+_18593097 | 3.26 |
ENST00000426208.1
|
TUBA8
|
tubulin, alpha 8 |
| chr21_+_30502806 | 3.21 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr10_-_75226166 | 3.05 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr5_+_54320078 | 2.99 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr9_-_71161505 | 2.93 |
ENST00000446290.1
|
RP11-274B18.2
|
RP11-274B18.2 |
| chr7_+_143013198 | 2.85 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr11_+_112832090 | 2.81 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr3_+_42190714 | 2.74 |
ENST00000449246.1
|
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr11_+_112832133 | 2.68 |
ENST00000524665.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr2_+_219247021 | 2.68 |
ENST00000539932.1
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chrY_+_2709527 | 2.43 |
ENST00000250784.8
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr7_-_36764004 | 2.34 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr6_-_130536774 | 2.32 |
ENST00000532763.1
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr7_-_36764062 | 2.16 |
ENST00000435386.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chrY_+_2709906 | 2.15 |
ENST00000430575.1
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr7_-_36764142 | 2.07 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr17_-_42452063 | 2.00 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr11_+_122709200 | 1.98 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr22_-_31688431 | 1.98 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr11_+_64073699 | 1.97 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr7_+_110731062 | 1.96 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chrX_-_19689106 | 1.91 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr22_-_31688381 | 1.90 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr11_+_61447845 | 1.84 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr7_-_142240014 | 1.83 |
ENST00000390363.2
|
TRBV9
|
T cell receptor beta variable 9 |
| chr2_-_40679186 | 1.82 |
ENST00000406785.2
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr1_-_11907829 | 1.81 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr22_+_40441456 | 1.81 |
ENST00000402203.1
|
TNRC6B
|
trinucleotide repeat containing 6B |
| chr1_-_25256368 | 1.77 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr6_-_160166218 | 1.71 |
ENST00000537657.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr1_-_154167124 | 1.70 |
ENST00000515609.1
|
TPM3
|
tropomyosin 3 |
| chr5_-_39270725 | 1.68 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr7_+_80253387 | 1.68 |
ENST00000438020.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_+_202995611 | 1.67 |
ENST00000367240.2
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr17_-_43502987 | 1.58 |
ENST00000376922.2
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr10_-_126432821 | 1.58 |
ENST00000280780.6
|
FAM53B
|
family with sequence similarity 53, member B |
| chr1_-_36947652 | 1.55 |
ENST00000533491.1
|
CSF3R
|
colony stimulating factor 3 receptor (granulocyte) |
| chr14_+_77582905 | 1.55 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chrX_+_47050798 | 1.53 |
ENST00000412206.1
ENST00000427561.1 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr12_+_69742121 | 1.53 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr12_-_90024360 | 1.50 |
ENST00000393164.2
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr1_-_205290865 | 1.48 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr15_-_38852251 | 1.47 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr12_-_89746264 | 1.47 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr12_-_89746173 | 1.44 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr1_+_172628154 | 1.42 |
ENST00000340030.3
ENST00000367721.2 |
FASLG
|
Fas ligand (TNF superfamily, member 6) |
| chr17_-_72619783 | 1.42 |
ENST00000328630.3
|
CD300E
|
CD300e molecule |
| chr12_+_58148842 | 1.39 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chr5_-_131132658 | 1.37 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr8_+_1993152 | 1.36 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr3_-_66551351 | 1.36 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr6_+_167536230 | 1.35 |
ENST00000341935.5
ENST00000349984.4 |
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr10_+_135050908 | 1.34 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chr1_-_156786530 | 1.33 |
ENST00000368198.3
|
SH2D2A
|
SH2 domain containing 2A |
| chr22_+_40342819 | 1.33 |
ENST00000407075.3
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr1_-_156786634 | 1.32 |
ENST00000392306.2
ENST00000368199.3 |
SH2D2A
|
SH2 domain containing 2A |
| chrX_+_109245863 | 1.30 |
ENST00000372072.3
|
TMEM164
|
transmembrane protein 164 |
| chr3_-_66551397 | 1.28 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr7_-_142120321 | 1.25 |
ENST00000390377.1
|
TRBV7-7
|
T cell receptor beta variable 7-7 |
| chr8_-_101724989 | 1.23 |
ENST00000517403.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr10_-_76868931 | 1.22 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr4_-_90757364 | 1.20 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_+_73358690 | 1.19 |
ENST00000545798.1
ENST00000539157.1 ENST00000546251.1 ENST00000535582.1 ENST00000538227.1 ENST00000543524.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr12_+_65672702 | 1.19 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr11_+_59824060 | 1.18 |
ENST00000395032.2
ENST00000358152.2 |
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr17_-_72619869 | 1.17 |
ENST00000392619.1
ENST00000426295.2 |
CD300E
|
CD300e molecule |
| chr7_+_123241908 | 1.16 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr11_+_64950801 | 1.14 |
ENST00000526468.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr5_+_150040403 | 1.14 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chrX_+_123480375 | 1.14 |
ENST00000360027.4
|
SH2D1A
|
SH2 domain containing 1A |
| chrX_+_123480421 | 1.12 |
ENST00000477673.2
|
SH2D1A
|
SH2 domain containing 1A |
| chr11_-_133826852 | 1.10 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chrX_+_123480194 | 1.10 |
ENST00000371139.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr16_+_27413483 | 1.10 |
ENST00000337929.3
ENST00000564089.1 |
IL21R
|
interleukin 21 receptor |
| chr4_+_160188889 | 1.10 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr8_-_66754172 | 1.09 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr2_+_219246746 | 1.09 |
ENST00000233202.6
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr12_-_111180644 | 1.09 |
ENST00000551676.1
ENST00000550991.1 ENST00000335007.5 ENST00000340766.5 |
PPP1CC
|
protein phosphatase 1, catalytic subunit, gamma isozyme |
| chr7_-_44179972 | 1.08 |
ENST00000446581.1
|
MYL7
|
myosin, light chain 7, regulatory |
| chr8_-_143833918 | 1.08 |
ENST00000359228.3
|
LYPD2
|
LY6/PLAUR domain containing 2 |
| chr10_+_45406627 | 1.08 |
ENST00000389583.4
|
TMEM72
|
transmembrane protein 72 |
| chr17_-_9929581 | 1.06 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr5_-_74807418 | 1.06 |
ENST00000405807.4
ENST00000261415.7 |
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr2_+_87808725 | 1.06 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr3_+_101498269 | 1.06 |
ENST00000491511.2
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr3_+_50654821 | 1.05 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr12_+_10183271 | 1.03 |
ENST00000355819.1
|
CLEC9A
|
C-type lectin domain family 9, member A |
| chr15_+_40861487 | 1.03 |
ENST00000315616.7
ENST00000559271.1 |
RPUSD2
|
RNA pseudouridylate synthase domain containing 2 |
| chr6_-_13487825 | 1.03 |
ENST00000603223.1
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr17_+_34538310 | 1.02 |
ENST00000444414.1
ENST00000378350.4 ENST00000389068.5 ENST00000588929.1 ENST00000589079.1 ENST00000589336.1 ENST00000400702.4 ENST00000591167.1 ENST00000586598.1 ENST00000591637.1 ENST00000378352.4 ENST00000358756.5 |
CCL4L1
|
chemokine (C-C motif) ligand 4-like 1 |
| chr15_+_41913690 | 1.02 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr1_-_158300747 | 1.02 |
ENST00000451207.1
|
CD1B
|
CD1b molecule |
| chr5_-_49737184 | 1.01 |
ENST00000508934.1
ENST00000303221.5 |
EMB
|
embigin |
| chr13_+_76378305 | 1.01 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr1_+_203256898 | 1.00 |
ENST00000433008.1
|
RP11-134P9.3
|
RP11-134P9.3 |
| chr1_-_11024258 | 0.98 |
ENST00000418570.2
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr11_+_59824127 | 0.97 |
ENST00000278865.3
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr9_+_34990219 | 0.97 |
ENST00000541010.1
ENST00000454002.2 ENST00000545841.1 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr4_-_90756769 | 0.96 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr14_-_62217779 | 0.96 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr10_-_23003460 | 0.96 |
ENST00000376573.4
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr1_+_40840320 | 0.96 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr14_+_98602380 | 0.95 |
ENST00000557072.1
|
RP11-61O1.2
|
RP11-61O1.2 |
| chr1_+_154975110 | 0.95 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr16_+_50313426 | 0.95 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr17_-_47308128 | 0.95 |
ENST00000413580.1
ENST00000511066.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr2_+_201994042 | 0.94 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_+_202172848 | 0.94 |
ENST00000255432.7
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6 |
| chr17_-_36884451 | 0.94 |
ENST00000595377.1
|
AC006449.1
|
NS5ATP13TP1; Uncharacterized protein |
| chr21_-_31864275 | 0.93 |
ENST00000334063.4
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chr11_+_73358594 | 0.93 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr12_+_66582919 | 0.93 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr1_+_184356188 | 0.92 |
ENST00000235307.6
|
C1orf21
|
chromosome 1 open reading frame 21 |
| chr12_-_68553512 | 0.92 |
ENST00000229135.3
|
IFNG
|
interferon, gamma |
| chr19_+_12862604 | 0.92 |
ENST00000553030.1
|
BEST2
|
bestrophin 2 |
| chr2_-_113594279 | 0.92 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr12_+_14572070 | 0.92 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr13_-_30160925 | 0.91 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr1_+_76540386 | 0.91 |
ENST00000328299.3
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr10_-_65028938 | 0.91 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr11_+_71846764 | 0.91 |
ENST00000456237.1
ENST00000442948.2 ENST00000546166.1 |
FOLR3
|
folate receptor 3 (gamma) |
| chr6_+_45390222 | 0.90 |
ENST00000359524.5
|
RUNX2
|
runt-related transcription factor 2 |
| chr13_+_42031679 | 0.89 |
ENST00000379359.3
|
RGCC
|
regulator of cell cycle |
| chr14_+_22433675 | 0.89 |
ENST00000390442.3
|
TRAV12-3
|
T cell receptor alpha variable 12-3 |
| chr20_+_58630972 | 0.89 |
ENST00000313426.1
|
C20orf197
|
chromosome 20 open reading frame 197 |
| chr19_+_55235969 | 0.88 |
ENST00000402254.2
ENST00000538269.1 ENST00000541392.1 ENST00000291860.1 ENST00000396284.2 |
KIR3DL1
KIR3DL3
KIR2DL4
|
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 3 killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4 |
| chr16_+_6069072 | 0.87 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr4_-_5991545 | 0.86 |
ENST00000531445.1
|
C4orf50
|
chromosome 4 open reading frame 50 |
| chr12_+_11905413 | 0.86 |
ENST00000545027.1
|
ETV6
|
ets variant 6 |
| chr7_+_77469439 | 0.85 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr7_+_31726822 | 0.84 |
ENST00000409146.3
|
PPP1R17
|
protein phosphatase 1, regulatory subunit 17 |
| chr7_+_107110488 | 0.84 |
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22 |
| chr11_-_33913708 | 0.84 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr7_+_142012967 | 0.84 |
ENST00000390357.3
|
TRBV4-1
|
T cell receptor beta variable 4-1 |
| chr10_-_76868866 | 0.83 |
ENST00000607487.1
|
DUSP13
|
dual specificity phosphatase 13 |
| chr4_+_108746282 | 0.83 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr1_+_32716857 | 0.82 |
ENST00000482949.1
ENST00000495610.2 |
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr1_+_20915409 | 0.82 |
ENST00000375071.3
|
CDA
|
cytidine deaminase |
| chr3_-_44465475 | 0.81 |
ENST00000416124.1
|
LINC00694
|
long intergenic non-protein coding RNA 694 |
| chr13_+_32634993 | 0.81 |
ENST00000436046.1
|
FRY
|
furry homolog (Drosophila) |
| chr16_+_30386098 | 0.80 |
ENST00000322861.7
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr5_-_88180342 | 0.80 |
ENST00000502983.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr2_+_120687335 | 0.80 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr7_+_150264365 | 0.80 |
ENST00000255945.2
ENST00000461940.1 |
GIMAP4
|
GTPase, IMAP family member 4 |
| chr4_-_8073554 | 0.79 |
ENST00000510277.1
|
ABLIM2
|
actin binding LIM protein family, member 2 |
| chr3_+_107243204 | 0.79 |
ENST00000456817.1
ENST00000458458.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr17_-_76713100 | 0.79 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr4_+_154622652 | 0.79 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr1_+_158325684 | 0.79 |
ENST00000368162.2
|
CD1E
|
CD1e molecule |
| chr2_-_25391507 | 0.79 |
ENST00000380794.1
|
POMC
|
proopiomelanocortin |
| chr11_+_107799118 | 0.78 |
ENST00000320578.2
|
RAB39A
|
RAB39A, member RAS oncogene family |
| chr10_-_16563870 | 0.78 |
ENST00000298943.3
|
C1QL3
|
complement component 1, q subcomponent-like 3 |
| chr14_+_75746664 | 0.78 |
ENST00000557139.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr7_-_142224280 | 0.77 |
ENST00000390367.3
|
TRBV11-1
|
T cell receptor beta variable 11-1 |
| chr1_+_32716840 | 0.77 |
ENST00000336890.5
|
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr6_-_13487784 | 0.77 |
ENST00000379287.3
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr1_-_36020531 | 0.77 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr2_+_29001711 | 0.76 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr7_+_150264470 | 0.76 |
ENST00000479232.1
|
GIMAP4
|
GTPase, IMAP family member 4 |
| chr10_+_118305435 | 0.76 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr7_+_107220660 | 0.76 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr12_-_91348949 | 0.75 |
ENST00000358859.2
|
CCER1
|
coiled-coil glutamate-rich protein 1 |
| chr17_+_75084717 | 0.75 |
ENST00000561721.2
ENST00000589827.1 ENST00000392476.2 |
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr6_-_6225026 | 0.75 |
ENST00000445223.1
|
F13A1
|
coagulation factor XIII, A1 polypeptide |
| chr6_+_45389893 | 0.74 |
ENST00000371432.3
|
RUNX2
|
runt-related transcription factor 2 |
| chr5_+_94727048 | 0.74 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr17_-_38721711 | 0.73 |
ENST00000578085.1
ENST00000246657.2 |
CCR7
|
chemokine (C-C motif) receptor 7 |
| chr1_-_11042094 | 0.73 |
ENST00000377004.4
ENST00000377008.4 |
C1orf127
|
chromosome 1 open reading frame 127 |
| chr1_+_174843548 | 0.72 |
ENST00000478442.1
ENST00000465412.1 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr19_-_51875894 | 0.72 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr12_-_76879852 | 0.72 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr12_-_52685312 | 0.71 |
ENST00000327741.5
|
KRT81
|
keratin 81 |
| chr6_-_31619697 | 0.71 |
ENST00000434444.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr17_-_1553346 | 0.71 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr1_+_39456895 | 0.71 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr12_-_51740463 | 0.71 |
ENST00000293636.1
|
CELA1
|
chymotrypsin-like elastase family, member 1 |
| chr10_+_94451574 | 0.71 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr17_+_34431212 | 0.71 |
ENST00000394495.1
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr16_+_57702099 | 0.70 |
ENST00000333493.4
ENST00000327655.6 |
GPR97
|
G protein-coupled receptor 97 |
| chr11_-_85779971 | 0.69 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr5_+_95998070 | 0.69 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chr1_+_10057274 | 0.69 |
ENST00000294435.7
|
RBP7
|
retinol binding protein 7, cellular |
| chr3_-_61237050 | 0.69 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr7_+_133261209 | 0.68 |
ENST00000545148.1
|
EXOC4
|
exocyst complex component 4 |
| chr3_+_171844762 | 0.68 |
ENST00000443501.1
|
FNDC3B
|
fibronectin type III domain containing 3B |
| chr17_-_37557846 | 0.67 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr12_+_131438496 | 0.67 |
ENST00000543826.1
|
GPR133
|
G protein-coupled receptor 133 |
| chr21_-_16374688 | 0.67 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr19_+_859654 | 0.66 |
ENST00000592860.1
|
CFD
|
complement factor D (adipsin) |
| chr14_+_23009190 | 0.66 |
ENST00000390532.1
|
TRAJ5
|
T cell receptor alpha joining 5 |
| chr14_-_98444369 | 0.66 |
ENST00000554822.1
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr9_+_12695702 | 0.66 |
ENST00000381136.2
|
TYRP1
|
tyrosinase-related protein 1 |
| chr19_-_46526304 | 0.66 |
ENST00000008938.4
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr20_-_36661826 | 0.65 |
ENST00000373448.2
ENST00000373447.3 |
TTI1
|
TELO2 interacting protein 1 |
| chr2_+_201994208 | 0.65 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX21_TBR1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.8 | 3.9 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.7 | 4.2 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.6 | 6.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.6 | 1.8 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.6 | 2.8 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.5 | 2.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.5 | 2.0 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.4 | 2.2 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.4 | 4.7 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.4 | 1.5 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.4 | 1.8 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.4 | 1.8 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.4 | 1.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.3 | 1.7 | GO:0003070 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.3 | 1.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.3 | 0.8 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.3 | 1.9 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.3 | 1.4 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.3 | 0.8 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.3 | 0.8 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.3 | 2.6 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.3 | 0.3 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.2 | 3.7 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 0.7 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.2 | 1.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.2 | 2.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 0.7 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.2 | 0.7 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.2 | 1.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 0.9 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.2 | 0.7 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.2 | 0.7 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.2 | 0.7 | GO:0051714 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.2 | 7.1 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.2 | 0.6 | GO:0003163 | sinoatrial node development(GO:0003163) sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.2 | 2.7 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.2 | 0.5 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 | 2.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 1.5 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.2 | 0.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 1.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 0.9 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.2 | 1.7 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 1.8 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 1.2 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 1.0 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.8 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.4 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.5 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 3.3 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 1.1 | GO:1901295 | positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 1.6 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.4 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.1 | 1.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.5 | GO:0071288 | cellular response to mercury ion(GO:0071288) positive regulation of mononuclear cell migration(GO:0071677) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 1.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.9 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.8 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.5 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.6 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.1 | 0.7 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.3 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.3 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.9 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.7 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 1.5 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.1 | 0.8 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.1 | 0.5 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 1.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.8 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 1.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 1.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.4 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.1 | 0.8 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.3 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 1.0 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 0.2 | GO:0048925 | lateral line system development(GO:0048925) |
| 0.1 | 0.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.4 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 1.6 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 7.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.1 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.1 | 0.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.3 | GO:0098907 | protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 0.3 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.2 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 1.0 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.3 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.5 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.1 | 0.4 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.3 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 1.8 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.7 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.5 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 0.9 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 1.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.3 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 1.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.2 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.1 | 0.6 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.1 | 0.6 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 0.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.0 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.8 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 0.9 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.1 | 1.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.5 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.6 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 2.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 1.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 7.7 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.7 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 1.6 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.7 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.4 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.3 | GO:0061366 | negative regulation of growth hormone secretion(GO:0060125) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 2.0 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.7 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 1.6 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.2 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.0 | 0.2 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 4.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.7 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 2.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.3 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.4 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 1.1 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.9 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 2.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.5 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.4 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.5 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 1.0 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.3 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.0 | 0.7 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 1.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 1.1 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.6 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.1 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 1.7 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.3 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.3 | GO:0051828 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.4 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.3 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 1.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 1.2 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.6 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 1.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.4 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 1.1 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.0 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 1.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.5 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 0.1 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.9 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 3.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 2.6 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 1.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 1.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 1.6 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.2 | 0.8 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 6.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.5 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 2.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.7 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.6 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.5 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 3.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.9 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 1.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.1 | 1.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 4.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 1.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 5.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 1.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 1.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 4.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.9 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 2.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 2.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.7 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 2.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 3.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 2.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 10.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 3.8 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 4.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 2.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.8 | GO:0031983 | vesicle lumen(GO:0031983) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 2.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.6 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 1.3 | 3.9 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.3 | 3.8 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.7 | 4.2 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.6 | 5.3 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.4 | 2.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.4 | 2.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.3 | 2.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 1.6 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.3 | 1.8 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.2 | 0.5 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 1.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 2.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 1.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 2.9 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 1.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.6 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 6.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 1.8 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.2 | 1.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.2 | 1.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 1.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.2 | 1.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 0.7 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 0.7 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.2 | 0.6 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 7.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.9 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.1 | 0.4 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 0.9 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.7 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 2.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.5 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 1.4 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.5 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 2.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.8 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 3.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.3 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.8 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.9 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 9.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.5 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 0.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 3.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 2.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 1.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 4.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.2 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 7.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 1.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 1.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 2.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 1.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 2.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.9 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 2.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 1.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.0 | 1.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 1.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.3 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 11.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 2.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 4.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 3.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 3.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 4.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 2.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 4.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 3.0 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 6.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 3.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.1 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 4.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 6.0 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 3.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.2 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 1.0 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.5 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 3.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 2.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 3.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.0 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |