Project
Illumina Body Map 2
Navigation
Downloads
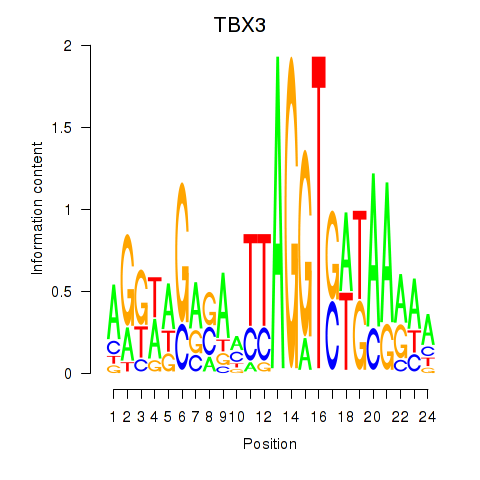
Results for TBX3
Z-value: 1.22
Transcription factors associated with TBX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX3
|
ENSG00000135111.10 | T-box transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX3 | hg19_v2_chr12_-_115121962_115121987 | -0.08 | 6.8e-01 | Click! |
Activity profile of TBX3 motif
Sorted Z-values of TBX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_156824840 | 4.44 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr6_-_52668605 | 3.83 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr20_+_56725952 | 3.73 |
ENST00000371168.3
|
C20orf85
|
chromosome 20 open reading frame 85 |
| chr13_-_46679144 | 3.43 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr22_+_22749343 | 3.26 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr2_-_89310012 | 3.18 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr5_+_54320078 | 3.00 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr14_-_106642049 | 2.98 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr3_+_149192475 | 2.93 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr22_+_23237555 | 2.90 |
ENST00000390321.2
|
IGLC1
|
immunoglobulin lambda constant 1 (Mcg marker) |
| chr2_-_89266286 | 2.88 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr14_-_106471723 | 2.73 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr16_+_72088376 | 2.72 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr14_+_22475742 | 2.63 |
ENST00000390447.3
|
TRAV19
|
T cell receptor alpha variable 19 |
| chr2_-_89513402 | 2.49 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr2_+_90229045 | 2.48 |
ENST00000390278.2
|
IGKV1D-42
|
immunoglobulin kappa variable 1D-42 (non-functional) |
| chr3_+_149191723 | 2.36 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr22_+_23063100 | 2.35 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr4_+_155484155 | 2.24 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr14_-_94919103 | 2.16 |
ENST00000334708.3
|
SERPINA11
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr4_+_40195262 | 2.14 |
ENST00000503941.1
|
RHOH
|
ras homolog family member H |
| chr11_-_18258342 | 2.12 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr2_+_241807870 | 2.09 |
ENST00000307503.3
|
AGXT
|
alanine-glyoxylate aminotransferase |
| chr4_+_155484103 | 2.08 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr17_-_7017559 | 1.99 |
ENST00000446679.2
|
ASGR2
|
asialoglycoprotein receptor 2 |
| chr14_-_107283278 | 1.91 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr4_-_71532207 | 1.87 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr7_+_142448053 | 1.86 |
ENST00000422143.2
|
TRBV29-1
|
T cell receptor beta variable 29-1 |
| chr3_+_108541545 | 1.83 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr2_+_89184868 | 1.82 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr4_-_84030996 | 1.77 |
ENST00000411416.2
|
PLAC8
|
placenta-specific 8 |
| chr6_-_52628271 | 1.73 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr2_+_90153696 | 1.70 |
ENST00000417279.2
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr3_+_108541608 | 1.65 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr14_+_22433675 | 1.63 |
ENST00000390442.3
|
TRAV12-3
|
T cell receptor alpha variable 12-3 |
| chr15_+_38296918 | 1.63 |
ENST00000557883.2
|
RP11-1008C21.1
|
RP11-1008C21.1 |
| chr2_+_90259593 | 1.61 |
ENST00000471857.1
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr2_+_90192768 | 1.59 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr22_+_22723969 | 1.57 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr5_-_80689945 | 1.56 |
ENST00000307624.3
|
ACOT12
|
acyl-CoA thioesterase 12 |
| chr2_-_209118974 | 1.54 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr2_+_96331830 | 1.49 |
ENST00000425887.2
|
AC008268.1
|
AC008268.1 |
| chr12_+_11081828 | 1.48 |
ENST00000381847.3
ENST00000396400.3 |
PRH2
|
proline-rich protein HaeIII subfamily 2 |
| chr2_-_89545079 | 1.45 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr11_-_104893863 | 1.45 |
ENST00000260315.3
ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5
|
caspase 5, apoptosis-related cysteine peptidase |
| chr19_-_3478477 | 1.41 |
ENST00000591708.1
|
C19orf77
|
chromosome 19 open reading frame 77 |
| chr1_+_159772121 | 1.36 |
ENST00000339348.5
ENST00000392235.3 ENST00000368106.3 |
FCRL6
|
Fc receptor-like 6 |
| chr7_-_99381884 | 1.34 |
ENST00000336411.2
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chrX_-_131623043 | 1.33 |
ENST00000421707.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr9_-_113100088 | 1.33 |
ENST00000374510.4
ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa) |
| chr19_-_33360647 | 1.33 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr11_+_7506837 | 1.32 |
ENST00000528758.1
|
OLFML1
|
olfactomedin-like 1 |
| chr3_+_52454971 | 1.30 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr3_-_42452050 | 1.28 |
ENST00000441172.1
ENST00000287748.3 |
LYZL4
|
lysozyme-like 4 |
| chrX_+_73524020 | 1.24 |
ENST00000339534.2
|
ZCCHC13
|
zinc finger, CCHC domain containing 13 |
| chr10_+_71561704 | 1.24 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr1_+_94798754 | 1.23 |
ENST00000418242.1
|
RP11-148B18.3
|
RP11-148B18.3 |
| chr20_+_29956369 | 1.23 |
ENST00000253381.2
|
DEFB118
|
defensin, beta 118 |
| chr21_-_15918618 | 1.22 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr1_+_212208919 | 1.21 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr22_+_24999114 | 1.19 |
ENST00000412658.1
ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr2_-_209119831 | 1.17 |
ENST00000345146.2
|
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr19_-_51530916 | 1.17 |
ENST00000594768.1
|
KLK11
|
kallikrein-related peptidase 11 |
| chr17_-_55822653 | 1.15 |
ENST00000299415.2
|
AC007431.1
|
coiled-coil domain containing 182 |
| chr4_+_187148556 | 1.14 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr2_-_133429091 | 1.13 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr1_+_162351503 | 1.13 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr2_+_234580499 | 1.11 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr10_+_71561630 | 1.11 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr19_-_51531272 | 1.11 |
ENST00000319720.7
|
KLK11
|
kallikrein-related peptidase 11 |
| chr11_-_104916034 | 1.09 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr19_-_52005009 | 1.08 |
ENST00000291707.3
|
SIGLEC12
|
sialic acid binding Ig-like lectin 12 (gene/pseudogene) |
| chr1_+_172628154 | 1.06 |
ENST00000340030.3
ENST00000367721.2 |
FASLG
|
Fas ligand (TNF superfamily, member 6) |
| chr2_-_4021609 | 1.06 |
ENST00000425949.1
ENST00000453880.1 ENST00000588796.1 |
AC107070.1
|
AC107070.1 |
| chr19_+_1077393 | 1.05 |
ENST00000590577.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr4_+_15704573 | 1.05 |
ENST00000265016.4
|
BST1
|
bone marrow stromal cell antigen 1 |
| chr11_+_7506713 | 1.05 |
ENST00000329293.3
ENST00000534244.1 |
OLFML1
|
olfactomedin-like 1 |
| chr6_+_126661253 | 1.04 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr7_-_99381798 | 1.02 |
ENST00000415003.1
ENST00000354593.2 |
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr6_-_32821599 | 1.01 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr22_+_24891210 | 1.00 |
ENST00000382760.2
|
UPB1
|
ureidopropionase, beta |
| chr6_+_160542821 | 1.00 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr19_-_7766991 | 0.98 |
ENST00000597921.1
ENST00000346664.5 |
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23) |
| chr1_+_159770292 | 0.98 |
ENST00000536257.1
ENST00000321935.6 |
FCRL6
|
Fc receptor-like 6 |
| chr20_+_55108302 | 0.98 |
ENST00000371325.1
|
FAM209B
|
family with sequence similarity 209, member B |
| chr2_+_234580525 | 0.98 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr7_+_22766766 | 0.98 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr1_-_182921119 | 0.97 |
ENST00000423786.1
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like |
| chr11_+_45743858 | 0.97 |
ENST00000532307.1
|
CTD-2210P24.1
|
CTD-2210P24.1 |
| chrX_+_150884539 | 0.96 |
ENST00000417321.1
|
FATE1
|
fetal and adult testis expressed 1 |
| chr4_-_46911223 | 0.96 |
ENST00000396533.1
|
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr16_+_27078219 | 0.94 |
ENST00000418886.1
|
C16orf82
|
chromosome 16 open reading frame 82 |
| chrX_-_101726732 | 0.93 |
ENST00000457521.2
ENST00000412230.2 ENST00000453326.2 |
NXF2B
TCP11X2
|
nuclear RNA export factor 2B t-complex 11 family, X-linked 2 |
| chr7_-_93519471 | 0.93 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr14_+_24641820 | 0.93 |
ENST00000560501.1
|
REC8
|
REC8 meiotic recombination protein |
| chr19_-_51531210 | 0.92 |
ENST00000391804.3
|
KLK11
|
kallikrein-related peptidase 11 |
| chr14_-_21056121 | 0.92 |
ENST00000557105.1
ENST00000398008.2 ENST00000555841.1 ENST00000443456.2 ENST00000432835.2 ENST00000557503.1 ENST00000398009.2 ENST00000554842.1 |
RNASE11
|
ribonuclease, RNase A family, 11 (non-active) |
| chr7_+_137761199 | 0.92 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr12_+_7169887 | 0.90 |
ENST00000542978.1
|
C1S
|
complement component 1, s subcomponent |
| chr21_-_46340807 | 0.90 |
ENST00000397846.3
ENST00000524251.1 ENST00000522688.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr17_-_60883993 | 0.90 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr11_+_34643600 | 0.90 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr17_-_3375033 | 0.90 |
ENST00000268981.5
ENST00000397168.3 ENST00000572969.1 ENST00000355380.4 ENST00000571553.1 ENST00000574797.1 ENST00000575375.1 |
SPATA22
|
spermatogenesis associated 22 |
| chr6_+_32821924 | 0.89 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr3_+_133118839 | 0.89 |
ENST00000302334.2
|
BFSP2
|
beaded filament structural protein 2, phakinin |
| chr10_+_71561649 | 0.87 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chrX_+_101470280 | 0.86 |
ENST00000395088.2
ENST00000330252.5 ENST00000333110.5 |
NXF2
TCP11X1
|
nuclear RNA export factor 2 t-complex 11 family, X-linked 1 |
| chr12_+_12223867 | 0.85 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr12_+_57849048 | 0.85 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chr17_+_45286387 | 0.85 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr11_+_77774897 | 0.84 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chrX_-_70838306 | 0.84 |
ENST00000373691.4
ENST00000373693.3 |
CXCR3
|
chemokine (C-X-C motif) receptor 3 |
| chr7_+_117119187 | 0.84 |
ENST00000446805.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr1_+_94713418 | 0.83 |
ENST00000413103.1
|
RP11-148B18.1
|
RP11-148B18.1 |
| chr2_-_4021643 | 0.82 |
ENST00000454455.1
|
AC107070.1
|
AC107070.1 |
| chr11_-_10590118 | 0.81 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr17_+_20483037 | 0.79 |
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2 |
| chr10_-_44274273 | 0.79 |
ENST00000419406.1
|
RP11-272J7.4
|
RP11-272J7.4 |
| chr19_-_51538148 | 0.79 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr11_+_18433840 | 0.76 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr15_-_50411412 | 0.76 |
ENST00000284509.6
|
ATP8B4
|
ATPase, class I, type 8B, member 4 |
| chr11_-_327537 | 0.75 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr2_+_149974684 | 0.74 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr4_+_123300664 | 0.74 |
ENST00000388725.2
|
ADAD1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr2_+_234637754 | 0.73 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr8_-_121825088 | 0.72 |
ENST00000520717.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr19_+_9361606 | 0.72 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr10_+_90750378 | 0.71 |
ENST00000355740.2
ENST00000352159.4 |
FAS
|
Fas cell surface death receptor |
| chr11_-_10590238 | 0.70 |
ENST00000256178.3
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr6_+_6588316 | 0.70 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr2_+_202937972 | 0.69 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr20_+_123010 | 0.69 |
ENST00000382398.3
|
DEFB126
|
defensin, beta 126 |
| chr16_+_63764128 | 0.68 |
ENST00000563061.1
|
RP11-370P15.2
|
RP11-370P15.2 |
| chr3_-_172019686 | 0.68 |
ENST00000596321.1
|
AC092964.2
|
Uncharacterized protein |
| chr9_-_100954910 | 0.68 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr14_+_90422867 | 0.67 |
ENST00000553989.1
|
TDP1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr15_-_38852251 | 0.66 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr11_+_45743931 | 0.66 |
ENST00000530051.1
|
CTD-2210P24.1
|
CTD-2210P24.1 |
| chr12_-_7848364 | 0.66 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3 |
| chr2_-_55459485 | 0.66 |
ENST00000451916.1
|
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr20_+_2795626 | 0.65 |
ENST00000603872.1
ENST00000380589.4 |
C20orf141
|
chromosome 20 open reading frame 141 |
| chr9_+_273038 | 0.64 |
ENST00000487230.1
ENST00000469391.1 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr20_-_1472053 | 0.64 |
ENST00000537284.1
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr16_-_67360662 | 0.63 |
ENST00000304372.5
|
KCTD19
|
potassium channel tetramerization domain containing 19 |
| chrX_-_131352152 | 0.62 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr4_-_80247162 | 0.62 |
ENST00000286794.4
|
NAA11
|
N(alpha)-acetyltransferase 11, NatA catalytic subunit |
| chr1_-_89488510 | 0.62 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr11_-_104972158 | 0.62 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr1_+_151020175 | 0.61 |
ENST00000368926.5
|
C1orf56
|
chromosome 1 open reading frame 56 |
| chr8_+_23386305 | 0.60 |
ENST00000519973.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr19_-_43835582 | 0.60 |
ENST00000595748.1
|
CTC-490G23.2
|
CTC-490G23.2 |
| chr2_+_162101247 | 0.58 |
ENST00000439050.1
ENST00000436506.1 |
AC009299.3
|
AC009299.3 |
| chr7_+_150434430 | 0.58 |
ENST00000358647.3
|
GIMAP5
|
GTPase, IMAP family member 5 |
| chr2_-_37415690 | 0.57 |
ENST00000260637.3
ENST00000535679.1 ENST00000379149.2 |
SULT6B1
|
sulfotransferase family, cytosolic, 6B, member 1 |
| chr1_-_183560011 | 0.57 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr4_-_46911248 | 0.56 |
ENST00000355591.3
ENST00000505102.1 |
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr12_+_1800179 | 0.56 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chrX_+_44703249 | 0.55 |
ENST00000339042.4
|
DUSP21
|
dual specificity phosphatase 21 |
| chr19_+_41103063 | 0.55 |
ENST00000308370.7
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr19_-_51538118 | 0.55 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr19_+_51754526 | 0.54 |
ENST00000596117.1
|
SIGLECL1
|
SIGLEC family like 1 |
| chr22_-_39928823 | 0.53 |
ENST00000334678.3
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr17_-_72619783 | 0.52 |
ENST00000328630.3
|
CD300E
|
CD300e molecule |
| chr2_-_165698662 | 0.52 |
ENST00000194871.6
ENST00000445474.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr20_-_1472029 | 0.51 |
ENST00000359801.3
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr7_-_150721570 | 0.51 |
ENST00000377974.2
ENST00000444312.1 ENST00000605938.1 ENST00000605952.1 |
ATG9B
|
autophagy related 9B |
| chr14_+_22958441 | 0.51 |
ENST00000390488.1
|
TRAJ49
|
T cell receptor alpha joining 49 |
| chr11_+_125616184 | 0.50 |
ENST00000305738.5
ENST00000437148.2 |
PATE1
|
prostate and testis expressed 1 |
| chr4_+_178230985 | 0.50 |
ENST00000264596.3
|
NEIL3
|
nei endonuclease VIII-like 3 (E. coli) |
| chr3_+_118866222 | 0.50 |
ENST00000490594.1
|
RP11-484M3.5
|
Uncharacterized protein |
| chr2_+_114647617 | 0.50 |
ENST00000536059.1
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast) |
| chr19_-_47291843 | 0.49 |
ENST00000542575.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr5_-_39425222 | 0.48 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr2_+_55459808 | 0.48 |
ENST00000404735.1
|
RPS27A
|
ribosomal protein S27a |
| chr5_-_174871136 | 0.48 |
ENST00000393752.2
|
DRD1
|
dopamine receptor D1 |
| chr4_+_103790462 | 0.47 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr9_-_130679257 | 0.46 |
ENST00000361444.3
ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr2_+_66666432 | 0.46 |
ENST00000495021.2
|
MEIS1
|
Meis homeobox 1 |
| chr11_+_71249071 | 0.46 |
ENST00000398534.3
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr2_-_24149977 | 0.45 |
ENST00000238789.5
|
ATAD2B
|
ATPase family, AAA domain containing 2B |
| chr7_-_112726393 | 0.45 |
ENST00000449591.1
ENST00000449735.1 ENST00000438062.1 ENST00000424100.1 |
GPR85
|
G protein-coupled receptor 85 |
| chr19_+_32516768 | 0.45 |
ENST00000593056.1
ENST00000588694.1 ENST00000585739.1 |
CTC-360P9.3
|
CTC-360P9.3 |
| chr6_-_35888905 | 0.44 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr1_-_119869846 | 0.44 |
ENST00000457719.1
|
RP11-418J17.3
|
RP11-418J17.3 |
| chr19_+_31658405 | 0.43 |
ENST00000589511.1
|
CTC-439O9.3
|
CTC-439O9.3 |
| chr16_+_55512742 | 0.43 |
ENST00000568715.1
ENST00000219070.4 |
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr7_+_150264365 | 0.42 |
ENST00000255945.2
ENST00000461940.1 |
GIMAP4
|
GTPase, IMAP family member 4 |
| chr2_+_55459495 | 0.42 |
ENST00000272317.6
ENST00000449323.1 |
RPS27A
|
ribosomal protein S27a |
| chr22_+_27017921 | 0.42 |
ENST00000354760.3
|
CRYBA4
|
crystallin, beta A4 |
| chr1_+_171217622 | 0.40 |
ENST00000433267.1
ENST00000367750.3 |
FMO1
|
flavin containing monooxygenase 1 |
| chr20_-_1472121 | 0.40 |
ENST00000444444.2
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr8_+_86851932 | 0.39 |
ENST00000517368.1
|
CTA-392E5.1
|
CTA-392E5.1 |
| chr11_+_71259466 | 0.39 |
ENST00000528743.2
|
KRTAP5-9
|
keratin associated protein 5-9 |
| chr14_+_23009190 | 0.39 |
ENST00000390532.1
|
TRAJ5
|
T cell receptor alpha joining 5 |
| chr14_-_23446900 | 0.38 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chr6_-_46703092 | 0.38 |
ENST00000541026.1
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr7_+_99971129 | 0.38 |
ENST00000394000.2
ENST00000350573.2 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chrX_-_65259914 | 0.37 |
ENST00000374737.4
ENST00000455586.2 |
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chrX_-_65259900 | 0.37 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr19_-_55677920 | 0.37 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.9 | 2.7 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.9 | 2.7 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.9 | 3.4 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.7 | 2.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.6 | 2.4 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.4 | 3.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 1.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.3 | 1.7 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.3 | 4.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 0.8 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.3 | 1.8 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.3 | 1.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.3 | 1.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.2 | 1.0 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.2 | 0.9 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.2 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.2 | 0.6 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.2 | 0.8 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.2 | 0.6 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.2 | 1.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.2 | 1.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 | 1.7 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.2 | 1.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.2 | 0.9 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.2 | 1.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.2 | 0.6 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 | 1.4 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 1.0 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.1 | 5.6 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 1.2 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 10.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.5 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 0.3 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.1 | 0.7 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 1.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 22.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.7 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 0.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 0.3 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.5 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.5 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.7 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 1.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.7 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 1.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 0.5 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 8.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.1 | 0.4 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.9 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.3 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.1 | 0.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.4 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.7 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 1.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.6 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 1.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 3.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 1.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 5.3 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.9 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.8 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0043449 | coumarin metabolic process(GO:0009804) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.5 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 2.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.4 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 1.2 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.8 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 1.1 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 2.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 2.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 1.1 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 2.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.9 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.6 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.5 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.2 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.8 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.6 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.7 | 2.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 4.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 2.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.2 | 1.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.2 | 1.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 10.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 0.9 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.8 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.9 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 1.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.9 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.7 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 0.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 3.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 3.2 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 4.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.0 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.9 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0016939 | plus-end kinesin complex(GO:0005873) kinesin II complex(GO:0016939) |
| 0.0 | 2.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 2.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 4.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 4.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 21.8 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.7 | 4.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.6 | 2.4 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.5 | 2.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 1.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.3 | 1.6 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.3 | 2.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.3 | 1.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 1.0 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.2 | 2.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.2 | 0.8 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.2 | 0.6 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.2 | 0.9 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.2 | 0.8 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.2 | 10.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.2 | 0.6 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 1.0 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.7 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 1.7 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.5 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 3.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.1 | 3.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 23.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.7 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 1.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 1.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 1.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 1.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 3.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.9 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 2.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 3.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 0.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 1.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.5 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 1.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.7 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.4 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 10.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 4.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.7 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.2 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.6 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 3.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 4.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 6.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 3.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 2.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 3.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |