Project
Illumina Body Map 2
Navigation
Downloads
Results for TEAD3_TEAD1
Z-value: 4.56
Transcription factors associated with TEAD3_TEAD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
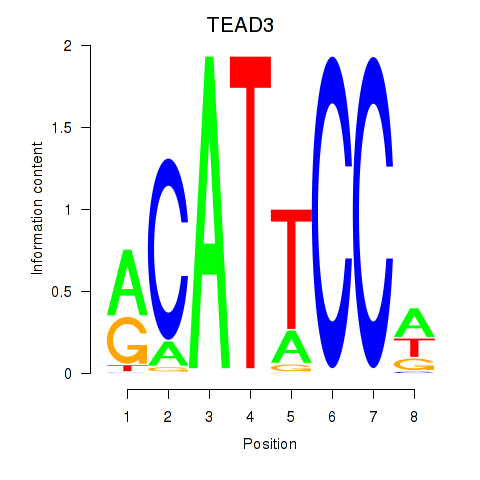
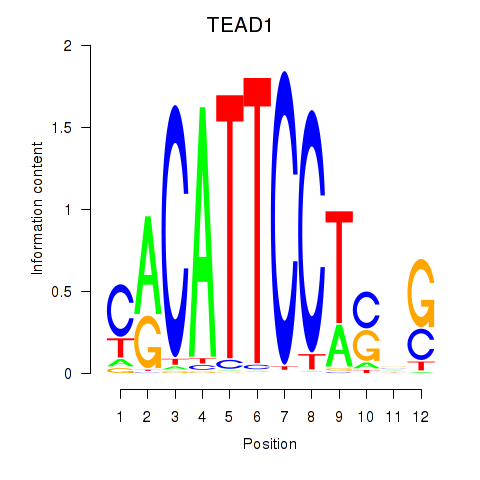
TEAD3
|
ENSG00000007866.14 | TEA domain transcription factor 3 |
|
TEAD1
|
ENSG00000187079.10 | TEA domain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD1 | hg19_v2_chr11_+_12766583_12766592 | 0.90 | 2.1e-12 | Click! |
| TEAD3 | hg19_v2_chr6_-_35464817_35464894 | 0.79 | 8.3e-08 | Click! |
Activity profile of TEAD3_TEAD1 motif
Sorted Z-values of TEAD3_TEAD1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_132272504 | 33.59 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr1_-_16344500 | 27.53 |
ENST00000406363.2
ENST00000411503.1 ENST00000545268.1 ENST00000487046.1 |
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular) |
| chr3_-_119379427 | 26.48 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr7_+_128470431 | 25.86 |
ENST00000325888.8
ENST00000346177.6 |
FLNC
|
filamin C, gamma |
| chr3_-_119379719 | 23.38 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr14_-_23904861 | 23.19 |
ENST00000355349.3
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta |
| chr5_-_138842286 | 20.40 |
ENST00000515823.1
|
ECSCR
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr11_-_111781454 | 18.02 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr4_-_186733363 | 18.01 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_86046433 | 17.38 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr10_-_90712520 | 16.84 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr14_-_23877474 | 16.54 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr17_-_40575535 | 16.37 |
ENST00000357037.5
|
PTRF
|
polymerase I and transcript release factor |
| chr11_+_66824276 | 14.88 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr11_+_66824346 | 14.87 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr10_+_123872483 | 14.75 |
ENST00000369001.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr10_+_24498060 | 14.75 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr15_+_96869165 | 14.26 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr16_-_122619 | 14.08 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr10_+_24497704 | 13.96 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr12_-_7245125 | 13.81 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr3_-_112360116 | 13.51 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr11_+_66824303 | 13.49 |
ENST00000533360.1
|
RHOD
|
ras homolog family member D |
| chr1_-_201438282 | 13.26 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr4_-_187644930 | 13.06 |
ENST00000441802.2
|
FAT1
|
FAT atypical cadherin 1 |
| chr4_+_184020398 | 12.99 |
ENST00000403733.3
ENST00000378925.3 |
WWC2
|
WW and C2 domain containing 2 |
| chr6_-_35464817 | 12.96 |
ENST00000338863.7
|
TEAD3
|
TEA domain family member 3 |
| chr6_-_35464727 | 12.83 |
ENST00000402886.3
|
TEAD3
|
TEA domain family member 3 |
| chr1_-_26394114 | 12.69 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr12_-_7245152 | 12.50 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr6_+_118869452 | 12.45 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr9_+_72658490 | 12.34 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr6_+_30850697 | 12.03 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr4_-_186732892 | 11.91 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_-_7245018 | 11.88 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr17_-_27949911 | 11.62 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr19_+_45409011 | 11.58 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr12_-_96184533 | 11.56 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr11_-_111782484 | 11.50 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr15_+_63334831 | 11.33 |
ENST00000288398.6
ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1
|
tropomyosin 1 (alpha) |
| chr12_-_96184913 | 11.31 |
ENST00000538383.1
|
NTN4
|
netrin 4 |
| chr2_-_217560248 | 11.29 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr4_-_152149033 | 11.23 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr11_-_111782696 | 10.92 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr14_-_89021077 | 10.82 |
ENST00000556564.1
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr8_-_82395461 | 10.57 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr5_-_38595498 | 10.50 |
ENST00000263409.4
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr12_-_7245080 | 10.48 |
ENST00000541042.1
ENST00000540242.1 |
C1R
|
complement component 1, r subcomponent |
| chr15_+_62853562 | 10.48 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr1_-_1293904 | 10.24 |
ENST00000309212.6
ENST00000342753.4 ENST00000445648.2 |
MXRA8
|
matrix-remodelling associated 8 |
| chr11_-_111781554 | 10.06 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr2_+_27505260 | 9.99 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr1_+_78354330 | 9.99 |
ENST00000440324.1
|
NEXN
|
nexilin (F actin binding protein) |
| chr16_+_30387141 | 9.95 |
ENST00000566955.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr11_-_111781610 | 9.94 |
ENST00000525823.1
|
CRYAB
|
crystallin, alpha B |
| chr3_-_134093275 | 9.93 |
ENST00000513145.1
ENST00000422605.2 |
AMOTL2
|
angiomotin like 2 |
| chr7_+_116165754 | 9.69 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr1_-_201346761 | 9.67 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr1_+_78354175 | 9.55 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr7_+_116165038 | 9.53 |
ENST00000393470.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr19_+_15218180 | 9.50 |
ENST00000342784.2
ENST00000597977.1 ENST00000600440.1 |
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr10_-_17659234 | 9.44 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr3_-_134093395 | 9.35 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr1_+_78354243 | 9.34 |
ENST00000294624.8
|
NEXN
|
nexilin (F actin binding protein) |
| chr19_+_34972543 | 9.13 |
ENST00000590071.2
|
WTIP
|
Wilms tumor 1 interacting protein |
| chr1_-_229569834 | 9.02 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr1_-_94050668 | 9.01 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_-_201915590 | 8.93 |
ENST00000367288.4
|
LMOD1
|
leiomodin 1 (smooth muscle) |
| chr2_+_228029281 | 8.89 |
ENST00000396578.3
|
COL4A3
|
collagen, type IV, alpha 3 (Goodpasture antigen) |
| chr17_-_39677971 | 8.88 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr8_+_70378852 | 8.87 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chrX_-_46187069 | 8.70 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr1_-_11918988 | 8.69 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr7_+_134464376 | 8.60 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr4_-_186733119 | 8.49 |
ENST00000419063.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_134464414 | 8.45 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr1_+_78354297 | 8.42 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr22_+_26138108 | 8.39 |
ENST00000536101.1
ENST00000335473.7 ENST00000407587.2 |
MYO18B
|
myosin XVIIIB |
| chr4_+_41614720 | 8.25 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr11_+_114166536 | 8.25 |
ENST00000299964.3
|
NNMT
|
nicotinamide N-methyltransferase |
| chr1_+_183155373 | 8.21 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr9_-_13175823 | 8.17 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr16_+_28889703 | 8.11 |
ENST00000357084.3
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr19_+_50691437 | 8.07 |
ENST00000598205.1
|
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr1_-_95391315 | 8.05 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr4_+_41614909 | 8.01 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_-_24438664 | 7.94 |
ENST00000374434.3
ENST00000330966.7 ENST00000329601.7 |
MYOM3
|
myomesin 3 |
| chr16_+_28889801 | 7.89 |
ENST00000395503.4
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr18_+_32173276 | 7.86 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr11_+_111782934 | 7.80 |
ENST00000304298.3
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr6_-_75915757 | 7.73 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr16_+_30386098 | 7.71 |
ENST00000322861.7
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr12_-_47219733 | 7.66 |
ENST00000547477.1
ENST00000447411.1 ENST00000266579.4 |
SLC38A4
|
solute carrier family 38, member 4 |
| chr5_+_145316120 | 7.65 |
ENST00000359120.4
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr14_-_23451845 | 7.65 |
ENST00000262713.2
|
AJUBA
|
ajuba LIM protein |
| chr15_-_99548775 | 7.56 |
ENST00000378919.6
|
PGPEP1L
|
pyroglutamyl-peptidase I-like |
| chr11_-_8832521 | 7.55 |
ENST00000530438.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr16_-_46797149 | 7.50 |
ENST00000536476.1
|
MYLK3
|
myosin light chain kinase 3 |
| chr3_+_9944303 | 7.44 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr3_-_134093738 | 7.44 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2 |
| chr12_-_8815404 | 7.42 |
ENST00000359478.2
ENST00000396549.2 |
MFAP5
|
microfibrillar associated protein 5 |
| chr1_-_59043166 | 7.35 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr10_+_69869237 | 7.29 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr12_+_27677085 | 7.28 |
ENST00000545334.1
ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr12_-_8815215 | 7.28 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr4_-_186877502 | 7.15 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_-_111358372 | 7.15 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr6_+_53883790 | 6.97 |
ENST00000509997.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr22_+_41956767 | 6.97 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr11_-_47470703 | 6.91 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chrX_+_99899180 | 6.89 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr4_+_55096489 | 6.86 |
ENST00000504461.1
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr19_-_54984354 | 6.85 |
ENST00000301200.2
|
CDC42EP5
|
CDC42 effector protein (Rho GTPase binding) 5 |
| chr10_+_112404132 | 6.76 |
ENST00000369519.3
|
RBM20
|
RNA binding motif protein 20 |
| chr14_+_32964258 | 6.62 |
ENST00000556638.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr12_+_3068466 | 6.61 |
ENST00000358409.2
|
TEAD4
|
TEA domain family member 4 |
| chr14_+_63671105 | 6.57 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr11_-_76381029 | 6.57 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr2_-_211179883 | 6.43 |
ENST00000352451.3
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr12_+_3068544 | 6.42 |
ENST00000540314.1
ENST00000536826.1 ENST00000359864.2 |
TEAD4
|
TEA domain family member 4 |
| chr7_+_134430212 | 6.30 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr11_+_46299199 | 6.20 |
ENST00000529193.1
ENST00000288400.3 |
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr2_-_218770168 | 6.19 |
ENST00000413554.1
|
TNS1
|
tensin 1 |
| chr20_-_62680984 | 6.15 |
ENST00000340356.7
|
SOX18
|
SRY (sex determining region Y)-box 18 |
| chr11_-_119249805 | 6.15 |
ENST00000527843.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr4_-_186732048 | 6.14 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_+_70117153 | 6.10 |
ENST00000245479.2
|
SOX9
|
SRY (sex determining region Y)-box 9 |
| chr11_-_47470682 | 6.04 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
| chr17_+_4901199 | 6.03 |
ENST00000320785.5
ENST00000574165.1 |
KIF1C
|
kinesin family member 1C |
| chr19_-_49567124 | 6.02 |
ENST00000301411.3
|
NTF4
|
neurotrophin 4 |
| chr10_+_50507232 | 5.99 |
ENST00000374144.3
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr19_-_46272462 | 5.98 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr12_-_8815299 | 5.97 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr16_+_23194033 | 5.95 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr10_-_97175444 | 5.95 |
ENST00000486141.2
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr6_+_53883708 | 5.93 |
ENST00000514921.1
ENST00000274897.5 ENST00000370877.2 |
MLIP
|
muscular LMNA-interacting protein |
| chr16_-_46782221 | 5.85 |
ENST00000394809.4
|
MYLK3
|
myosin light chain kinase 3 |
| chr4_+_169552748 | 5.85 |
ENST00000504519.1
ENST00000512127.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_+_114522049 | 5.83 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr12_-_12419905 | 5.83 |
ENST00000535731.1
|
LRP6
|
low density lipoprotein receptor-related protein 6 |
| chr4_+_166300084 | 5.82 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr2_-_216257849 | 5.77 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr4_+_110834033 | 5.77 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr3_-_149375783 | 5.75 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr10_+_123923205 | 5.70 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chrX_-_10851762 | 5.69 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr22_+_24577183 | 5.67 |
ENST00000358321.3
|
SUSD2
|
sushi domain containing 2 |
| chr21_-_40033618 | 5.63 |
ENST00000417133.2
ENST00000398910.1 ENST00000442448.1 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr5_+_42423872 | 5.57 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr21_+_17566643 | 5.54 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr6_+_19837592 | 5.53 |
ENST00000378700.3
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr2_+_170366203 | 5.52 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr2_+_189157536 | 5.50 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr13_-_33760216 | 5.46 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr10_+_81891416 | 5.45 |
ENST00000372270.2
|
PLAC9
|
placenta-specific 9 |
| chr9_+_27109440 | 5.45 |
ENST00000519080.1
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr17_-_73511504 | 5.43 |
ENST00000581870.1
|
CASKIN2
|
CASK interacting protein 2 |
| chr11_-_8832182 | 5.41 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr11_-_68780824 | 5.41 |
ENST00000441623.1
ENST00000309099.6 |
MRGPRF
|
MAS-related GPR, member F |
| chr10_+_50507181 | 5.40 |
ENST00000323868.4
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr1_-_161279749 | 5.37 |
ENST00000533357.1
ENST00000360451.6 ENST00000336559.4 |
MPZ
|
myelin protein zero |
| chr3_-_52868931 | 5.31 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr9_-_14308004 | 5.31 |
ENST00000493697.1
|
NFIB
|
nuclear factor I/B |
| chr9_+_109694914 | 5.30 |
ENST00000542028.1
|
ZNF462
|
zinc finger protein 462 |
| chr4_-_186877806 | 5.30 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_-_73511584 | 5.29 |
ENST00000321617.3
|
CASKIN2
|
CASK interacting protein 2 |
| chr2_+_189157498 | 5.28 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr9_+_27109392 | 5.28 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr1_+_178310581 | 5.22 |
ENST00000462775.1
|
RASAL2
|
RAS protein activator like 2 |
| chr1_+_66999799 | 5.20 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr5_-_58882219 | 5.19 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr14_+_63671577 | 5.18 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr3_+_9944492 | 5.18 |
ENST00000383814.3
ENST00000454190.2 ENST00000454992.1 |
IL17RE
|
interleukin 17 receptor E |
| chr15_-_48937982 | 5.11 |
ENST00000316623.5
|
FBN1
|
fibrillin 1 |
| chr17_+_73717516 | 5.06 |
ENST00000200181.3
ENST00000339591.3 |
ITGB4
|
integrin, beta 4 |
| chr11_-_47470591 | 5.01 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chr3_-_66024213 | 4.97 |
ENST00000483466.1
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chrX_-_33229636 | 4.96 |
ENST00000357033.4
|
DMD
|
dystrophin |
| chr12_-_8815477 | 4.92 |
ENST00000433590.2
|
MFAP5
|
microfibrillar associated protein 5 |
| chr11_+_101981169 | 4.92 |
ENST00000526343.1
ENST00000282441.5 ENST00000537274.1 ENST00000345877.2 |
YAP1
|
Yes-associated protein 1 |
| chr15_+_65369082 | 4.92 |
ENST00000432196.2
|
KBTBD13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr16_+_30907927 | 4.90 |
ENST00000279804.2
ENST00000395019.3 |
CTF1
|
cardiotrophin 1 |
| chr6_-_55740352 | 4.83 |
ENST00000370830.3
|
BMP5
|
bone morphogenetic protein 5 |
| chr14_-_23446900 | 4.81 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chr1_+_62417957 | 4.72 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr9_+_113431029 | 4.72 |
ENST00000189978.5
ENST00000374448.4 ENST00000374440.3 |
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr11_+_7506713 | 4.71 |
ENST00000329293.3
ENST00000534244.1 |
OLFML1
|
olfactomedin-like 1 |
| chr9_+_102584128 | 4.69 |
ENST00000338488.4
ENST00000395097.2 |
NR4A3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr3_-_149051444 | 4.68 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr1_+_19967014 | 4.67 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr4_+_111397216 | 4.66 |
ENST00000265162.5
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A) |
| chr5_-_149516966 | 4.66 |
ENST00000517957.1
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr5_+_150406527 | 4.62 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr19_-_49371711 | 4.62 |
ENST00000355496.5
ENST00000263265.6 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr17_+_73717407 | 4.62 |
ENST00000579662.1
|
ITGB4
|
integrin, beta 4 |
| chr5_-_41213607 | 4.61 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr8_-_81787006 | 4.56 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chr15_+_74466744 | 4.56 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr3_-_149051194 | 4.55 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr4_+_55096010 | 4.55 |
ENST00000503856.1
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD3_TEAD1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.2 | 33.6 | GO:0034059 | response to anoxia(GO:0034059) |
| 9.8 | 39.2 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 6.4 | 19.2 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 6.0 | 18.0 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 4.3 | 25.9 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 4.1 | 12.4 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 3.9 | 11.6 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) positive regulation of receptor catabolic process(GO:2000646) |
| 3.8 | 22.9 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 3.8 | 11.3 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 3.6 | 14.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 3.3 | 13.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 3.2 | 9.7 | GO:0035261 | external genitalia morphogenesis(GO:0035261) canonical Wnt signaling pathway involved in neural crest cell differentiation(GO:0044335) |
| 3.1 | 6.1 | GO:0072197 | ureter urothelium development(GO:0072190) ureter morphogenesis(GO:0072197) |
| 2.7 | 13.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 2.5 | 17.8 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 2.5 | 12.7 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 2.5 | 35.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 2.5 | 19.9 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 2.4 | 60.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 2.4 | 12.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 2.4 | 9.5 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 2.4 | 61.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 2.2 | 17.4 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 2.1 | 4.2 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 2.1 | 16.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 2.0 | 6.0 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 2.0 | 25.8 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 2.0 | 5.9 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 1.9 | 9.7 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 1.9 | 5.8 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 1.9 | 9.3 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 1.8 | 14.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.8 | 12.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.7 | 10.3 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 1.6 | 13.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.6 | 1.6 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 1.6 | 6.5 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 1.6 | 3.3 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 1.6 | 4.7 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 1.5 | 6.0 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 1.4 | 21.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.4 | 4.2 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 1.4 | 70.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 1.3 | 4.0 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 1.3 | 4.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 1.3 | 3.9 | GO:0048320 | axial mesoderm formation(GO:0048320) negative regulation of lymphangiogenesis(GO:1901491) |
| 1.2 | 6.2 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 1.2 | 9.8 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 1.2 | 9.5 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 1.2 | 5.8 | GO:0030070 | insulin processing(GO:0030070) |
| 1.1 | 12.6 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 1.1 | 6.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.1 | 14.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 1.0 | 1.0 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 1.0 | 25.0 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 1.0 | 13.0 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.0 | 9.9 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 1.0 | 56.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 1.0 | 6.9 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 1.0 | 6.8 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.9 | 6.6 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.9 | 4.6 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.9 | 5.5 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.9 | 8.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.9 | 4.5 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.8 | 2.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.8 | 14.2 | GO:0035878 | nail development(GO:0035878) |
| 0.8 | 8.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.8 | 5.6 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.8 | 5.6 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.8 | 8.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.8 | 6.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.8 | 1.5 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.7 | 6.7 | GO:0072093 | metanephric renal vesicle formation(GO:0072093) |
| 0.7 | 18.8 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.7 | 1.4 | GO:0002035 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.7 | 12.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.7 | 11.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.7 | 4.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.7 | 4.6 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.6 | 3.1 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.6 | 0.6 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.6 | 6.8 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.6 | 3.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.6 | 4.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.6 | 38.5 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.6 | 4.7 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.6 | 2.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.6 | 9.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.6 | 11.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.6 | 2.8 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.6 | 3.3 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.5 | 1.6 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.5 | 10.2 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.5 | 25.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.5 | 10.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.5 | 3.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.5 | 35.6 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.5 | 5.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.5 | 11.8 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.5 | 7.9 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.5 | 1.8 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.5 | 6.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.5 | 2.3 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.4 | 1.8 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 3.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.4 | 1.3 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.4 | 2.6 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.4 | 5.2 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.4 | 1.3 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.4 | 8.2 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.4 | 8.6 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.4 | 1.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.4 | 9.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.4 | 1.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.4 | 2.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.4 | 8.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.4 | 1.2 | GO:1904888 | cranial skeletal system development(GO:1904888) |
| 0.4 | 52.0 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.4 | 2.3 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.4 | 11.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 3.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.4 | 2.9 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.4 | 4.6 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.4 | 0.7 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.3 | 54.1 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.3 | 5.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.3 | 6.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 3.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 2.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 0.9 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.3 | 11.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 2.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.3 | 6.7 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.3 | 1.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 1.5 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.3 | 4.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 7.9 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.3 | 1.7 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.3 | 0.3 | GO:2000630 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) positive regulation of miRNA metabolic process(GO:2000630) |
| 0.3 | 1.4 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.3 | 3.6 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 3.0 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.3 | 3.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 1.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.3 | 1.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.3 | 8.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 1.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 8.8 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.2 | 5.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 3.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.7 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 1.4 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.2 | 3.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 0.2 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.2 | 0.7 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 1.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 0.6 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.2 | 2.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.2 | 2.8 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.2 | 2.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 3.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 4.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.2 | 1.0 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.2 | 0.6 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.2 | 5.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 6.0 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.2 | 37.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.2 | 3.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 7.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 3.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 4.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 1.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 4.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 2.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 | 2.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 1.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 3.7 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.2 | 2.3 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 2.4 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 1.8 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 2.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 0.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 0.5 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.2 | 7.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 1.7 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 2.6 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 | 7.5 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 5.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 13.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.7 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.4 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 4.5 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 20.1 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 3.6 | GO:0010842 | retina layer formation(GO:0010842) retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 3.1 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 0.9 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.7 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 1.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 6.8 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 18.9 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 5.3 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 0.3 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 1.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 19.8 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.1 | 26.2 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 1.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 5.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.9 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 0.3 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.1 | 1.3 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 0.5 | GO:0061046 | regulation of branching involved in lung morphogenesis(GO:0061046) |
| 0.1 | 0.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 1.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.7 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.5 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.6 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 1.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 3.0 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 4.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 11.1 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.1 | 0.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 1.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.0 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.4 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 2.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.8 | GO:0060526 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.1 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 1.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 14.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 1.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 3.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.5 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.0 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.1 | 1.0 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.6 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 1.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 3.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 4.8 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 2.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 3.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 0.4 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 6.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 4.2 | GO:0051782 | negative regulation of cell division(GO:0051782) |
| 0.0 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 1.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 1.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 1.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.4 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.0 | 2.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 1.2 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 3.8 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 1.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 2.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 3.1 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 1.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.6 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 1.7 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.5 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 0.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 2.6 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 1.8 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.3 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 1.8 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 1.2 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 9.1 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.6 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.3 | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.8 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 8.8 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.3 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.9 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 1.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 1.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.8 | GO:1903363 | negative regulation of cellular protein catabolic process(GO:1903363) |
| 0.0 | 2.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 78.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.9 | 11.6 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 3.0 | 8.9 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 2.8 | 16.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 2.7 | 8.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 2.6 | 7.7 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 2.1 | 8.2 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 2.0 | 52.0 | GO:0032982 | myosin filament(GO:0032982) |
| 1.9 | 9.7 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.9 | 5.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.8 | 3.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 1.4 | 35.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.4 | 4.2 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 1.2 | 18.0 | GO:0030478 | actin cap(GO:0030478) |
| 1.2 | 4.8 | GO:0097182 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 1.1 | 28.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 1.1 | 13.0 | GO:0016013 | syntrophin complex(GO:0016013) |
| 1.1 | 18.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.1 | 18.0 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 1.0 | 11.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.0 | 8.9 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 1.0 | 4.9 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 1.0 | 14.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.9 | 6.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.8 | 2.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 3.0 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.7 | 42.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.7 | 22.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.7 | 178.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.7 | 14.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.7 | 13.1 | GO:0032059 | bleb(GO:0032059) |
| 0.6 | 8.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.6 | 8.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.5 | 22.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.5 | 1.5 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.5 | 1.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.5 | 4.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 4.2 | GO:0031674 | I band(GO:0031674) |
| 0.4 | 3.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.4 | 17.9 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.4 | 31.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.4 | 29.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 1.1 | GO:0000806 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) |
| 0.4 | 2.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.3 | 8.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 0.9 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 3.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 6.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.3 | 3.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 4.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 52.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.3 | 30.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 6.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 7.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 7.9 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 2.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 51.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 1.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.2 | 1.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 8.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 4.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 32.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 1.6 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 2.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 14.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 3.9 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 84.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 14.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 10.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 2.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 12.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 10.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 2.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 10.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 5.9 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 40.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 1.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 10.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 7.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 37.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 11.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 19.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 4.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 40.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 11.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 2.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 4.4 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 6.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 2.8 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 5.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.2 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 3.2 | 9.7 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 3.2 | 19.4 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 2.9 | 11.6 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 2.9 | 11.4 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 2.2 | 20.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 2.1 | 8.2 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 2.0 | 17.6 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 1.8 | 56.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.8 | 12.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.7 | 75.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 1.6 | 4.9 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 1.6 | 7.9 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 1.5 | 63.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.4 | 12.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.4 | 4.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.4 | 4.2 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 1.4 | 30.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 1.4 | 45.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.3 | 13.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.3 | 12.7 | GO:0031432 | titin binding(GO:0031432) |
| 1.2 | 12.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.2 | 4.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 1.1 | 121.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 1.1 | 12.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.1 | 31.7 | GO:0031005 | filamin binding(GO:0031005) |
| 1.1 | 16.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.0 | 7.7 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.9 | 3.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.9 | 1.8 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.9 | 6.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.9 | 29.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.9 | 6.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.9 | 8.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.9 | 13.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.8 | 24.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.8 | 4.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.8 | 6.4 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.8 | 3.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.7 | 8.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.7 | 7.7 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.7 | 6.7 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.7 | 13.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.6 | 2.5 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.6 | 1.8 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.6 | 7.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.6 | 1.7 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.5 | 4.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.5 | 11.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.5 | 13.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.5 | 4.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.5 | 2.9 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.5 | 9.9 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.4 | 13.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 4.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.4 | 5.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.4 | 13.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.4 | 5.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.4 | 9.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 1.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.4 | 14.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.4 | 1.2 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.4 | 3.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.4 | 6.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.4 | 14.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 2.6 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.4 | 5.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.4 | 1.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.3 | 1.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.3 | 2.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 1.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 1.0 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.3 | 4.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 3.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 3.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.3 | 2.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.3 | 2.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 1.2 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.3 | 23.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.3 | 1.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.3 | 10.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 3.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 9.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 3.0 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.3 | 1.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.3 | 7.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 5.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 4.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 27.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 6.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 4.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 2.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 2.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 0.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 1.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 0.6 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.2 | 5.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.2 | 17.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 5.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 1.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 13.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 1.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 0.7 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.2 | 10.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 0.8 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.2 | 3.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 6.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 1.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 40.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 6.4 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.2 | 1.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 0.9 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 8.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 2.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 19.8 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.2 | 4.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 2.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 4.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 28.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 17.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 4.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 3.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 3.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 3.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 56.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.9 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.7 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.6 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 7.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 2.0 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 13.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 36.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 5.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 39.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 5.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 11.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.6 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 1.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.3 | GO:0015563 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 15.1 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.1 | 1.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 16.0 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.1 | 35.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 2.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.5 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 4.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 2.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 1.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.8 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 3.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.4 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 1.0 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 4.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 5.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.5 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.7 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 4.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 8.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 8.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.6 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.9 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.6 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 25.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.7 | 28.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.6 | 24.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.5 | 29.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.5 | 19.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 12.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 3.9 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 12.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.3 | 7.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 30.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.3 | 33.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.3 | 4.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 25.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 31.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 10.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 21.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 17.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 17.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.2 | 10.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 17.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 4.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 5.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 6.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.2 | 6.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 13.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 6.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 2.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 19.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 5.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 5.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 6.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 5.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 23.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.6 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 2.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 16.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 5.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 10.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 82.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 1.1 | 144.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 1.0 | 36.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.9 | 33.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.7 | 19.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.6 | 25.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.6 | 3.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.6 | 25.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.6 | 16.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.6 | 14.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.5 | 15.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 18.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.4 | 5.8 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.4 | 3.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.4 | 16.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.4 | 1.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.3 | 28.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 4.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 37.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 19.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 5.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 5.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 6.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 6.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 3.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 10.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 8.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 4.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 15.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 7.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 4.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 30.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 10.7 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 2.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 6.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 8.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 2.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 4.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.3 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 1.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 4.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 5.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 10.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 2.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |