Project
Illumina Body Map 2
Navigation
Downloads
Results for TFAP4_MSC
Z-value: 2.05
Transcription factors associated with TFAP4_MSC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP4
|
ENSG00000090447.7 | transcription factor AP-4 |
|
MSC
|
ENSG00000178860.8 | musculin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSC | hg19_v2_chr8_-_72756667_72756736 | -0.52 | 2.3e-03 | Click! |
| TFAP4 | hg19_v2_chr16_-_4323015_4323076 | -0.14 | 4.5e-01 | Click! |
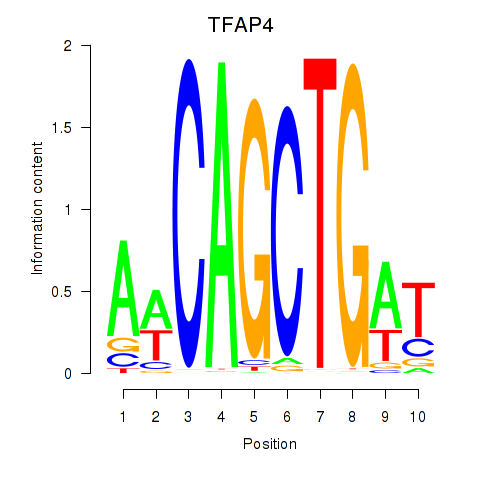
Activity profile of TFAP4_MSC motif
Sorted Z-values of TFAP4_MSC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_152590946 | 13.64 |
ENST00000172853.10
|
NEB
|
nebulin |
| chr1_+_167063282 | 13.43 |
ENST00000361200.2
|
DUSP27
|
dual specificity phosphatase 27 (putative) |
| chr14_+_32964258 | 10.84 |
ENST00000556638.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr3_+_8775466 | 10.13 |
ENST00000343849.2
ENST00000397368.2 |
CAV3
|
caveolin 3 |
| chr2_-_175629135 | 9.77 |
ENST00000409542.1
ENST00000409219.1 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr2_-_175629164 | 9.69 |
ENST00000409323.1
ENST00000261007.5 ENST00000348749.5 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr16_+_30383613 | 9.59 |
ENST00000568749.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr4_-_177190364 | 8.88 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr17_+_65040678 | 7.97 |
ENST00000226021.3
|
CACNG1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr2_+_27505260 | 7.50 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr8_+_86351056 | 7.09 |
ENST00000285381.2
|
CA3
|
carbonic anhydrase III, muscle specific |
| chr10_+_88428370 | 6.53 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr22_-_36013368 | 6.42 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr12_-_57644952 | 6.24 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr15_-_42749711 | 6.10 |
ENST00000565611.1
ENST00000263805.4 ENST00000565948.1 |
ZNF106
|
zinc finger protein 106 |
| chr16_+_7560114 | 5.68 |
ENST00000570626.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_-_179672142 | 5.66 |
ENST00000342992.6
ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN
|
titin |
| chr1_+_172422026 | 5.59 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr8_-_144512576 | 5.56 |
ENST00000333480.2
|
MAFA
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
| chr14_+_32963433 | 5.53 |
ENST00000554410.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr2_+_233404429 | 5.50 |
ENST00000389494.3
ENST00000389492.3 |
CHRNG
|
cholinergic receptor, nicotinic, gamma (muscle) |
| chrX_-_33357558 | 5.46 |
ENST00000288447.4
|
DMD
|
dystrophin |
| chr10_-_29923893 | 5.45 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr1_+_46379254 | 5.12 |
ENST00000372008.2
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr6_+_41021027 | 5.08 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr20_+_30407105 | 5.01 |
ENST00000375994.2
|
MYLK2
|
myosin light chain kinase 2 |
| chr20_+_30407151 | 5.01 |
ENST00000375985.4
|
MYLK2
|
myosin light chain kinase 2 |
| chr17_-_41132088 | 4.98 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr22_+_26138108 | 4.98 |
ENST00000536101.1
ENST00000335473.7 ENST00000407587.2 |
MYO18B
|
myosin XVIIIB |
| chr1_+_78354243 | 4.98 |
ENST00000294624.8
|
NEXN
|
nexilin (F actin binding protein) |
| chr10_+_88428206 | 4.93 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr3_-_46904946 | 4.73 |
ENST00000292327.4
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr3_-_46904918 | 4.71 |
ENST00000395869.1
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr1_+_78354330 | 4.69 |
ENST00000440324.1
|
NEXN
|
nexilin (F actin binding protein) |
| chr5_-_58571935 | 4.66 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr1_-_26394114 | 4.64 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr2_+_88367299 | 4.53 |
ENST00000419482.2
ENST00000444564.2 |
SMYD1
|
SET and MYND domain containing 1 |
| chr2_-_227050079 | 4.41 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr1_+_78354175 | 4.40 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr1_+_78354297 | 4.31 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr1_-_216896780 | 4.20 |
ENST00000459955.1
ENST00000366937.1 ENST00000408911.3 ENST00000391890.3 |
ESRRG
|
estrogen-related receptor gamma |
| chr8_-_72274095 | 4.20 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr1_-_201081579 | 4.13 |
ENST00000367338.3
ENST00000362061.3 |
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr1_+_203256898 | 4.08 |
ENST00000433008.1
|
RP11-134P9.3
|
RP11-134P9.3 |
| chr1_+_153600869 | 3.85 |
ENST00000292169.1
ENST00000368696.3 ENST00000436839.1 |
S100A1
|
S100 calcium binding protein A1 |
| chr2_+_88367368 | 3.85 |
ENST00000438570.1
|
SMYD1
|
SET and MYND domain containing 1 |
| chr2_-_152590982 | 3.78 |
ENST00000409198.1
ENST00000397345.3 ENST00000427231.2 |
NEB
|
nebulin |
| chr20_+_33292507 | 3.72 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr11_-_111783595 | 3.54 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B |
| chr5_+_191592 | 3.47 |
ENST00000328278.3
|
LRRC14B
|
leucine rich repeat containing 14B |
| chr17_+_45286387 | 3.39 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr11_+_1944054 | 3.39 |
ENST00000397301.1
ENST00000397304.2 ENST00000446240.1 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr16_+_83932684 | 3.33 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr17_-_62066769 | 3.33 |
ENST00000577329.1
|
CTC-264K15.6
|
CTC-264K15.6 |
| chr12_-_70093111 | 3.33 |
ENST00000548658.1
ENST00000476098.1 ENST00000331471.4 ENST00000393365.1 |
BEST3
|
bestrophin 3 |
| chr6_+_17281573 | 3.31 |
ENST00000379052.5
|
RBM24
|
RNA binding motif protein 24 |
| chr3_-_52486841 | 3.29 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr12_-_70093190 | 3.25 |
ENST00000330891.5
|
BEST3
|
bestrophin 3 |
| chr1_-_216896674 | 3.22 |
ENST00000475275.1
ENST00000469486.1 ENST00000481543.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr12_-_70093065 | 3.08 |
ENST00000553096.1
|
BEST3
|
bestrophin 3 |
| chr21_+_30673091 | 2.93 |
ENST00000447177.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr10_-_92681033 | 2.90 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr21_+_30672433 | 2.88 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr2_+_136343904 | 2.86 |
ENST00000436436.1
|
R3HDM1
|
R3H domain containing 1 |
| chr8_-_145016692 | 2.85 |
ENST00000357649.2
|
PLEC
|
plectin |
| chr2_+_233390863 | 2.83 |
ENST00000449596.1
ENST00000543200.1 |
CHRND
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr8_-_75233563 | 2.79 |
ENST00000342232.4
|
JPH1
|
junctophilin 1 |
| chr1_+_12042015 | 2.76 |
ENST00000412236.1
|
MFN2
|
mitofusin 2 |
| chr11_+_1860200 | 2.72 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr2_-_176867534 | 2.70 |
ENST00000445472.1
|
KIAA1715
|
KIAA1715 |
| chr3_+_42727011 | 2.64 |
ENST00000287777.4
|
KLHL40
|
kelch-like family member 40 |
| chr9_+_134165063 | 2.62 |
ENST00000372264.3
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr6_-_3912207 | 2.57 |
ENST00000566733.1
|
RP1-140K8.5
|
RP1-140K8.5 |
| chr8_-_72274355 | 2.57 |
ENST00000388741.2
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr18_+_7231123 | 2.56 |
ENST00000383467.2
|
LRRC30
|
leucine rich repeat containing 30 |
| chrX_+_135279179 | 2.53 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr12_-_54121261 | 2.52 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr17_+_67410832 | 2.50 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr13_+_41635617 | 2.49 |
ENST00000542082.1
|
WBP4
|
WW domain binding protein 4 |
| chr12_-_70093162 | 2.48 |
ENST00000551160.1
|
BEST3
|
bestrophin 3 |
| chr9_+_134165195 | 2.45 |
ENST00000372261.1
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr2_-_176867501 | 2.44 |
ENST00000535310.1
|
KIAA1715
|
KIAA1715 |
| chr5_+_150040403 | 2.40 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr8_-_72274467 | 2.39 |
ENST00000340726.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr5_+_102201509 | 2.37 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr5_+_102201687 | 2.36 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_-_81792780 | 2.34 |
ENST00000489715.1
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr15_+_86686953 | 2.30 |
ENST00000421325.2
|
AGBL1
|
ATP/GTP binding protein-like 1 |
| chr10_-_61899124 | 2.30 |
ENST00000373815.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr12_-_54121212 | 2.27 |
ENST00000548263.1
ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr6_-_125766306 | 2.27 |
ENST00000430672.1
|
RP11-138M12.1
|
RP11-138M12.1 |
| chr1_-_231560790 | 2.22 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr2_+_29341037 | 2.20 |
ENST00000449202.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr8_-_41522779 | 2.20 |
ENST00000522231.1
ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1
|
ankyrin 1, erythrocytic |
| chrX_+_70521584 | 2.12 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr5_+_102201722 | 2.12 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr6_-_110736742 | 2.12 |
ENST00000368924.3
ENST00000368923.3 |
DDO
|
D-aspartate oxidase |
| chr22_+_35695793 | 2.10 |
ENST00000456128.1
ENST00000449058.2 ENST00000411850.1 ENST00000425375.1 ENST00000436462.2 ENST00000382034.5 |
TOM1
|
target of myb1 (chicken) |
| chr3_-_52868931 | 2.09 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr2_+_136343820 | 2.08 |
ENST00000410054.1
|
R3HDM1
|
R3H domain containing 1 |
| chr2_+_33701707 | 2.08 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_-_154842741 | 2.04 |
ENST00000271915.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr5_+_102201430 | 2.02 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr19_+_35521616 | 2.01 |
ENST00000595652.1
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr2_-_176866978 | 2.01 |
ENST00000392540.2
ENST00000409660.1 ENST00000544803.1 ENST00000272748.4 |
KIAA1715
|
KIAA1715 |
| chr19_+_35521572 | 2.01 |
ENST00000262631.5
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chrX_+_135278908 | 1.99 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr4_-_153303658 | 1.95 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr9_-_123239632 | 1.93 |
ENST00000416449.1
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr1_+_10271674 | 1.93 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr20_+_33292068 | 1.93 |
ENST00000374810.3
ENST00000374809.2 ENST00000451665.1 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr17_+_45286706 | 1.91 |
ENST00000393450.1
ENST00000572303.1 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chrX_-_108868390 | 1.87 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr5_-_95158644 | 1.85 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr1_-_201346761 | 1.84 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr19_-_46285736 | 1.84 |
ENST00000291270.4
ENST00000447742.2 ENST00000354227.5 |
DMPK
|
dystrophia myotonica-protein kinase |
| chr1_-_33336414 | 1.84 |
ENST00000373471.3
ENST00000609187.1 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr3_+_32148106 | 1.83 |
ENST00000425459.1
ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr7_-_74867509 | 1.79 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr20_-_36152914 | 1.79 |
ENST00000397131.1
|
BLCAP
|
bladder cancer associated protein |
| chr3_+_32147997 | 1.78 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr19_+_35521699 | 1.77 |
ENST00000415950.3
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr10_-_97321112 | 1.77 |
ENST00000607232.1
ENST00000371227.4 ENST00000371249.2 ENST00000371247.2 ENST00000371246.2 ENST00000393949.1 ENST00000353505.5 ENST00000347291.4 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr6_+_155470243 | 1.77 |
ENST00000456877.2
ENST00000528391.2 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr5_+_43603229 | 1.73 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr10_+_13142075 | 1.72 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr7_+_73507409 | 1.72 |
ENST00000538333.3
|
LIMK1
|
LIM domain kinase 1 |
| chr11_+_34999328 | 1.71 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr17_-_27467418 | 1.70 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr8_+_9413410 | 1.68 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr8_-_123706338 | 1.68 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr1_-_154164534 | 1.67 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr10_+_13142225 | 1.67 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr14_+_90863327 | 1.67 |
ENST00000356978.4
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr6_-_24911195 | 1.64 |
ENST00000259698.4
|
FAM65B
|
family with sequence similarity 65, member B |
| chr7_+_80231466 | 1.63 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr3_-_52869205 | 1.63 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr17_+_73028676 | 1.62 |
ENST00000581589.1
|
KCTD2
|
potassium channel tetramerization domain containing 2 |
| chr17_+_42248063 | 1.60 |
ENST00000293414.1
|
ASB16
|
ankyrin repeat and SOCS box containing 16 |
| chr7_+_134832808 | 1.60 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr1_-_9129895 | 1.60 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr11_-_1785139 | 1.58 |
ENST00000236671.2
|
CTSD
|
cathepsin D |
| chr11_+_123430948 | 1.57 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr1_+_22328144 | 1.55 |
ENST00000290122.3
ENST00000374663.1 |
CELA3A
|
chymotrypsin-like elastase family, member 3A |
| chr8_+_98788057 | 1.55 |
ENST00000517924.1
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr2_+_33701286 | 1.55 |
ENST00000403687.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr10_-_97321165 | 1.54 |
ENST00000306402.6
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr5_+_176692466 | 1.54 |
ENST00000508029.1
ENST00000503056.1 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr12_+_652294 | 1.52 |
ENST00000322843.3
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr15_-_52944231 | 1.52 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr17_+_33570055 | 1.52 |
ENST00000299977.4
ENST00000542451.1 ENST00000592325.1 |
SLFN5
|
schlafen family member 5 |
| chr15_+_42696954 | 1.52 |
ENST00000337571.4
ENST00000569136.1 |
CAPN3
|
calpain 3, (p94) |
| chr19_-_46285646 | 1.51 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr7_+_80275953 | 1.51 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr10_+_24738355 | 1.51 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr15_+_42697018 | 1.49 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr12_-_70093235 | 1.49 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr17_+_4981535 | 1.48 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr9_-_35111420 | 1.46 |
ENST00000378557.1
|
FAM214B
|
family with sequence similarity 214, member B |
| chr8_-_66701319 | 1.45 |
ENST00000379419.4
|
PDE7A
|
phosphodiesterase 7A |
| chr3_+_49058444 | 1.45 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr4_+_77172847 | 1.45 |
ENST00000515604.1
ENST00000539752.1 ENST00000424749.2 |
FAM47E
FAM47E-STBD1
FAM47E
|
uncharacterized protein LOC100631383 FAM47E-STBD1 readthrough family with sequence similarity 47, member E |
| chr18_-_74728998 | 1.45 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr15_-_40401062 | 1.44 |
ENST00000354670.4
ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF
|
Bcl2 modifying factor |
| chr18_-_59274139 | 1.43 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr3_+_35721106 | 1.43 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_+_14075903 | 1.43 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr5_+_138209688 | 1.42 |
ENST00000518381.1
|
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr11_-_62323702 | 1.42 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr8_-_42396185 | 1.39 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr15_-_75748143 | 1.39 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr14_-_69619823 | 1.38 |
ENST00000341516.5
|
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr16_-_75150665 | 1.38 |
ENST00000300051.4
ENST00000450168.2 |
LDHD
|
lactate dehydrogenase D |
| chr3_+_69134080 | 1.36 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr16_+_85204882 | 1.35 |
ENST00000574293.1
|
CTC-786C10.1
|
CTC-786C10.1 |
| chr1_+_14075865 | 1.35 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr15_+_42697065 | 1.34 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr17_+_44679808 | 1.33 |
ENST00000571172.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr16_+_290181 | 1.32 |
ENST00000417499.1
|
ITFG3
|
integrin alpha FG-GAP repeat containing 3 |
| chr15_+_42696992 | 1.32 |
ENST00000561817.1
|
CAPN3
|
calpain 3, (p94) |
| chr6_+_155334780 | 1.31 |
ENST00000538270.1
ENST00000535231.1 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr14_+_24584508 | 1.31 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr19_+_10527449 | 1.31 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr9_+_125795788 | 1.29 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr13_+_76413852 | 1.29 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr20_+_53092232 | 1.26 |
ENST00000395939.1
|
DOK5
|
docking protein 5 |
| chr11_+_111783450 | 1.25 |
ENST00000537382.1
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr3_+_69134124 | 1.25 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr3_+_159570722 | 1.24 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr2_-_69870747 | 1.24 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr10_-_62149433 | 1.23 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chrX_+_105445717 | 1.23 |
ENST00000372552.1
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr17_+_46918925 | 1.22 |
ENST00000502761.1
|
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr2_+_136343943 | 1.22 |
ENST00000409606.1
|
R3HDM1
|
R3H domain containing 1 |
| chr10_+_94594351 | 1.22 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr11_-_1507965 | 1.22 |
ENST00000329957.6
|
MOB2
|
MOB kinase activator 2 |
| chr2_+_233562015 | 1.22 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr17_-_21156578 | 1.22 |
ENST00000399011.2
ENST00000468196.1 |
C17orf103
|
chromosome 17 open reading frame 103 |
| chr4_-_52904425 | 1.22 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr19_-_40324767 | 1.21 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP4_MSC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 17.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 2.5 | 22.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 2.2 | 15.2 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 2.0 | 16.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 2.0 | 15.8 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 1.5 | 8.9 | GO:0018032 | protein amidation(GO:0018032) |
| 1.4 | 5.8 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 1.3 | 6.5 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 1.2 | 3.6 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 1.2 | 4.6 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.9 | 18.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.8 | 9.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.8 | 6.4 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.7 | 2.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.7 | 4.9 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.7 | 3.3 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.7 | 2.0 | GO:1901076 | positive regulation of engulfment of apoptotic cell(GO:1901076) |
| 0.6 | 2.6 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.6 | 3.5 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.6 | 8.8 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.6 | 2.2 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.5 | 1.6 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.5 | 5.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.5 | 2.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.5 | 1.5 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.5 | 1.9 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.5 | 4.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.4 | 6.1 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.4 | 4.8 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.4 | 3.0 | GO:1990539 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.4 | 1.7 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.4 | 1.7 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.4 | 2.0 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.4 | 1.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.4 | 2.6 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.4 | 1.9 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.4 | 5.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.4 | 1.5 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.3 | 1.7 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.3 | 1.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.3 | 1.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 2.4 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.3 | 20.7 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.3 | 2.6 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.3 | 2.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 1.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 0.8 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.3 | 3.4 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.3 | 3.1 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.3 | 1.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 1.7 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.2 | 0.7 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 1.5 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.2 | 11.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 14.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.2 | 2.8 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.2 | 8.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.2 | 1.8 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.2 | 1.5 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.2 | 0.6 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 8.7 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.2 | 0.9 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 1.8 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 0.9 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.2 | 0.7 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 1.8 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.2 | 7.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 1.8 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 1.0 | GO:0014826 | cellular magnesium ion homeostasis(GO:0010961) vein smooth muscle contraction(GO:0014826) |
| 0.2 | 2.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 0.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.5 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.2 | 1.4 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 1.0 | GO:1902306 | negative regulation of sodium ion transmembrane transport(GO:1902306) |
| 0.1 | 3.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.4 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.1 | 1.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) chromatin maintenance(GO:0070827) |
| 0.1 | 1.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.5 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.1 | 0.5 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.4 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.8 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.1 | 0.6 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 1.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.8 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.8 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 1.1 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 | 7.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.5 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 1.9 | GO:0014870 | response to muscle inactivity(GO:0014870) |
| 0.1 | 1.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 1.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 2.2 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 2.3 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 1.0 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.5 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 1.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.8 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 1.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 2.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 7.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.6 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.3 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 0.4 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 1.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 2.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.5 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.2 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.1 | 2.5 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 1.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 2.4 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 2.8 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.3 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 0.5 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 0.3 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.8 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 1.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.3 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 1.0 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.6 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 1.0 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 0.6 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.2 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.1 | 0.5 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.1 | GO:0035284 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.5 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 10.3 | GO:0043271 | negative regulation of ion transport(GO:0043271) |
| 0.1 | 4.5 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 4.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 2.8 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.9 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 2.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.3 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 1.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:0050976 | sensory perception of touch(GO:0050975) detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.1 | 2.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 1.7 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 0.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.1 | 1.1 | GO:0090050 | peptidyl-lysine deacetylation(GO:0034983) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 3.7 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 2.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 2.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.5 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 2.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.7 | GO:0018210 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.1 | 0.8 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 0.5 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.1 | 2.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 8.0 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.2 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 3.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 1.2 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 1.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 2.7 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 1.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.3 | GO:1902022 | lysine transport(GO:0015819) L-lysine transport(GO:1902022) L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 1.1 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.3 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.4 | GO:0030638 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.6 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 1.8 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 3.1 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.3 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 1.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.6 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.5 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.2 | GO:0045345 | positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 2.8 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.4 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 1.7 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.2 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) |
| 0.0 | 0.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.5 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) |
| 0.0 | 0.4 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.6 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 1.7 | GO:0072413 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.8 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.3 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.5 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 4.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.4 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.8 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 1.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.0 | 0.4 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.1 | GO:0071440 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.7 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.5 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 1.1 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.1 | GO:0090678 | response to mycophenolic acid(GO:0071505) cellular response to mycophenolic acid(GO:0071506) metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.0 | 0.3 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 15.2 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.9 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.4 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.0 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.4 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.3 | GO:0000470 | maturation of 5.8S rRNA(GO:0000460) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 2.8 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.5 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.1 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 19.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.0 | 27.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.9 | 5.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.5 | 25.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.5 | 3.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.5 | 3.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.5 | 5.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.4 | 15.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 1.7 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.4 | 12.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 6.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 8.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.3 | 14.0 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 14.2 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 59.7 | GO:0031674 | I band(GO:0031674) |
| 0.3 | 7.9 | GO:0031672 | A band(GO:0031672) |
| 0.3 | 2.0 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 14.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.2 | 0.9 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.2 | 11.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 6.1 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.2 | 1.7 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 1.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.2 | 1.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 0.5 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) glial limiting end-foot(GO:0097451) |
| 0.1 | 2.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.7 | GO:0072357 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.9 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 1.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 3.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 2.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 1.0 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 6.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 3.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 3.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 8.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 7.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 9.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 1.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.5 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 1.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.3 | GO:0031310 | intrinsic component of endosome membrane(GO:0031302) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 1.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 14.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 6.7 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 3.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 1.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 8.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 3.4 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 1.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.4 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.5 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) cell-substrate junction(GO:0030055) |
| 0.0 | 1.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 6.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.1 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 15.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.7 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 1.5 | 8.9 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 1.4 | 5.8 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.2 | 3.6 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 1.1 | 27.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 1.1 | 10.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.0 | 2.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.9 | 7.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.8 | 3.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.8 | 10.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.8 | 16.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.7 | 5.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.6 | 1.7 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.6 | 5.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.6 | 9.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.5 | 8.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.5 | 63.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 2.0 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.5 | 4.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.5 | 1.4 | GO:0016898 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.4 | 1.3 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.4 | 3.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.4 | 7.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 1.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.3 | 1.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 1.2 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.3 | 6.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 6.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 3.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.3 | 1.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 1.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 2.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 0.7 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 4.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 2.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 1.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 1.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 0.8 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 1.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.7 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 5.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.9 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 4.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 2.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 10.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 13.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 0.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 1.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 2.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.4 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 2.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.9 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 2.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.4 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 0.8 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.8 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 8.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.0 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 3.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.7 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 1.6 | GO:0034594 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.1 | 0.6 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.4 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 3.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 3.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 3.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 2.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 3.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.0 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 2.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 9.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.6 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 1.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 4.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.4 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.2 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 2.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 2.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 1.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 11.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 9.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.6 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 6.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 4.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 5.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 1.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 4.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 11.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.6 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 1.9 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 4.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 3.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.5 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 3.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 1.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 9.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 1.1 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 13.9 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.2 | 16.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 4.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 4.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 3.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 2.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 9.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 4.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 5.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 2.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 2.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 3.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 27.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.6 | 45.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 7.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 12.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 6.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 4.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 6.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 3.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 3.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 1.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 6.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 21.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 6.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 1.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 2.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 4.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |