Project
Illumina Body Map 2
Navigation
Downloads
Results for THRA_RXRB
Z-value: 1.62
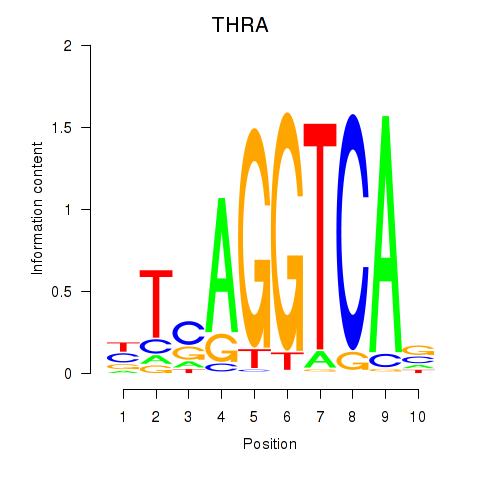
Transcription factors associated with THRA_RXRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRA
|
ENSG00000126351.8 | thyroid hormone receptor alpha |
|
RXRB
|
ENSG00000204231.6 | retinoid X receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RXRB | hg19_v2_chr6_-_33168391_33168465 | 0.23 | 2.0e-01 | Click! |
| THRA | hg19_v2_chr17_+_38219063_38219154 | 0.08 | 6.5e-01 | Click! |
Activity profile of THRA_RXRB motif
Sorted Z-values of THRA_RXRB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_150693318 | 11.49 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr1_-_150693305 | 7.36 |
ENST00000368987.1
|
HORMAD1
|
HORMA domain containing 1 |
| chr1_+_178482212 | 4.81 |
ENST00000319416.2
ENST00000258298.2 ENST00000367643.3 ENST00000367642.3 |
TEX35
|
testis expressed 35 |
| chr11_-_26593677 | 4.58 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr12_+_57854274 | 4.15 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr7_+_128431444 | 3.92 |
ENST00000459946.1
ENST00000378685.4 ENST00000464832.1 ENST00000472049.1 ENST00000488925.1 |
CCDC136
|
coiled-coil domain containing 136 |
| chr11_-_123756334 | 3.82 |
ENST00000528595.1
ENST00000375026.2 |
TMEM225
|
transmembrane protein 225 |
| chr19_-_55874611 | 3.80 |
ENST00000424985.3
|
FAM71E2
|
family with sequence similarity 71, member E2 |
| chrX_-_70326455 | 3.77 |
ENST00000374251.5
|
CXorf65
|
chromosome X open reading frame 65 |
| chr15_+_45422131 | 3.73 |
ENST00000321429.4
|
DUOX1
|
dual oxidase 1 |
| chr22_+_23264766 | 3.68 |
ENST00000390331.2
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chr5_+_162887556 | 3.63 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr12_-_52995291 | 3.57 |
ENST00000293745.2
ENST00000354310.4 |
KRT72
|
keratin 72 |
| chrX_-_30327495 | 3.53 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr20_-_13971255 | 3.50 |
ENST00000284951.5
ENST00000378072.5 |
SEL1L2
|
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr15_+_45422178 | 3.47 |
ENST00000389037.3
ENST00000558322.1 |
DUOX1
|
dual oxidase 1 |
| chr16_+_4784273 | 3.43 |
ENST00000299320.5
ENST00000586724.1 |
C16orf71
|
chromosome 16 open reading frame 71 |
| chr7_+_100187196 | 3.43 |
ENST00000468962.1
ENST00000427939.2 |
FBXO24
|
F-box protein 24 |
| chr12_+_7072354 | 3.13 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr15_-_45422056 | 3.11 |
ENST00000267803.4
ENST00000559014.1 ENST00000558851.1 ENST00000559988.1 ENST00000558996.1 ENST00000558422.1 ENST00000559226.1 ENST00000558326.1 ENST00000558377.1 ENST00000559644.1 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr19_-_35719609 | 3.04 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187, member B |
| chr19_+_49109990 | 3.02 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr16_+_4784458 | 2.98 |
ENST00000590191.1
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr22_+_51176624 | 2.94 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr1_+_160147176 | 2.78 |
ENST00000470705.1
|
ATP1A4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr19_-_52227221 | 2.71 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr20_+_32250079 | 2.68 |
ENST00000375222.3
|
C20orf144
|
chromosome 20 open reading frame 144 |
| chr16_-_11367452 | 2.66 |
ENST00000327157.2
|
PRM3
|
protamine 3 |
| chr1_-_38218577 | 2.61 |
ENST00000540011.1
|
EPHA10
|
EPH receptor A10 |
| chr20_+_32254286 | 2.60 |
ENST00000330271.4
|
ACTL10
|
actin-like 10 |
| chr2_-_55459485 | 2.50 |
ENST00000451916.1
|
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr9_-_127358087 | 2.48 |
ENST00000475178.1
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr1_+_212797789 | 2.47 |
ENST00000294829.3
|
FAM71A
|
family with sequence similarity 71, member A |
| chr13_+_113030625 | 2.44 |
ENST00000283550.3
|
SPACA7
|
sperm acrosome associated 7 |
| chr2_+_232457569 | 2.42 |
ENST00000313965.2
|
C2orf57
|
chromosome 2 open reading frame 57 |
| chr20_-_31592113 | 2.41 |
ENST00000420875.1
ENST00000375519.2 ENST00000375523.3 ENST00000356173.3 |
SUN5
|
Sad1 and UNC84 domain containing 5 |
| chr16_-_58328923 | 2.38 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr22_-_29137771 | 2.36 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr1_+_20396649 | 2.31 |
ENST00000375108.3
|
PLA2G5
|
phospholipase A2, group V |
| chr3_+_137490748 | 2.30 |
ENST00000478772.1
|
RP11-2A4.3
|
RP11-2A4.3 |
| chr22_+_22764088 | 2.30 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr7_+_73245193 | 2.28 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr18_-_53804580 | 2.28 |
ENST00000590484.1
ENST00000589293.1 ENST00000587904.1 ENST00000591974.1 |
RP11-456O19.4
|
RP11-456O19.4 |
| chr16_-_58328884 | 2.28 |
ENST00000569079.1
|
PRSS54
|
protease, serine, 54 |
| chr7_-_121944491 | 2.27 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr11_-_5531215 | 2.18 |
ENST00000311659.4
|
UBQLN3
|
ubiquilin 3 |
| chr11_+_66624527 | 2.18 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr16_-_72206034 | 2.16 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr8_+_103563792 | 2.13 |
ENST00000285402.3
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr14_-_106054659 | 2.11 |
ENST00000390539.2
|
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr2_-_55459437 | 2.10 |
ENST00000401408.1
|
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr12_-_5352315 | 2.10 |
ENST00000536518.1
|
RP11-319E16.1
|
RP11-319E16.1 |
| chr22_+_22697537 | 2.04 |
ENST00000427632.2
|
IGLV9-49
|
immunoglobulin lambda variable 9-49 |
| chr9_+_140145713 | 1.99 |
ENST00000388931.3
ENST00000412566.1 |
C9orf173
|
chromosome 9 open reading frame 173 |
| chr1_+_178482262 | 1.96 |
ENST00000367641.3
ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr16_-_58328870 | 1.96 |
ENST00000543437.1
|
PRSS54
|
protease, serine, 54 |
| chr9_+_35042205 | 1.95 |
ENST00000312292.5
ENST00000378745.3 |
C9orf131
|
chromosome 9 open reading frame 131 |
| chr2_+_230787213 | 1.91 |
ENST00000409992.1
|
FBXO36
|
F-box protein 36 |
| chr12_+_122018697 | 1.88 |
ENST00000541574.1
|
RP13-941N14.1
|
RP13-941N14.1 |
| chr3_-_126277796 | 1.87 |
ENST00000318225.2
|
C3orf22
|
chromosome 3 open reading frame 22 |
| chr22_+_22681656 | 1.86 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr2_+_220491973 | 1.85 |
ENST00000358055.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr7_+_120969045 | 1.76 |
ENST00000222462.2
|
WNT16
|
wingless-type MMTV integration site family, member 16 |
| chr11_-_5531159 | 1.75 |
ENST00000445998.1
|
UBQLN3
|
ubiquilin 3 |
| chr2_+_231860830 | 1.72 |
ENST00000424440.1
ENST00000452881.1 ENST00000433428.2 ENST00000455816.1 ENST00000440792.1 ENST00000423134.1 |
SPATA3
|
spermatogenesis associated 3 |
| chr3_+_10068095 | 1.72 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr1_-_203144941 | 1.69 |
ENST00000255416.4
|
MYBPH
|
myosin binding protein H |
| chr1_+_32827759 | 1.68 |
ENST00000373534.3
|
TSSK3
|
testis-specific serine kinase 3 |
| chr20_+_43343886 | 1.62 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr2_+_230787201 | 1.62 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr22_+_23165153 | 1.61 |
ENST00000390317.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr22_+_23213658 | 1.60 |
ENST00000390318.2
|
IGLV4-3
|
immunoglobulin lambda variable 4-3 |
| chr1_+_96457545 | 1.60 |
ENST00000413825.2
|
RP11-147C23.1
|
Uncharacterized protein |
| chr17_+_58499844 | 1.60 |
ENST00000269127.4
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr18_+_19749386 | 1.59 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr9_+_36572851 | 1.59 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr22_+_22516550 | 1.57 |
ENST00000390284.2
|
IGLV4-60
|
immunoglobulin lambda variable 4-60 |
| chr17_+_43922241 | 1.57 |
ENST00000329196.5
|
SPPL2C
|
signal peptide peptidase like 2C |
| chr8_-_30706608 | 1.55 |
ENST00000256246.2
|
TEX15
|
testis expressed 15 |
| chr7_+_26591441 | 1.52 |
ENST00000420912.1
ENST00000457000.1 ENST00000430426.1 |
AC004947.2
|
AC004947.2 |
| chr2_-_55459294 | 1.51 |
ENST00000407122.1
ENST00000406437.2 |
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr2_-_61389168 | 1.50 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr1_+_45212074 | 1.50 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr5_+_71014990 | 1.50 |
ENST00000296777.4
|
CARTPT
|
CART prepropeptide |
| chr17_+_60457914 | 1.48 |
ENST00000305286.3
ENST00000520404.1 ENST00000518576.1 |
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr12_+_102514019 | 1.47 |
ENST00000537257.1
ENST00000358383.5 ENST00000392911.2 |
PARPBP
|
PARP1 binding protein |
| chr2_+_219745020 | 1.46 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr7_+_23749945 | 1.45 |
ENST00000354639.3
ENST00000531170.1 ENST00000444333.2 ENST00000428484.1 |
STK31
|
serine/threonine kinase 31 |
| chr8_+_145086574 | 1.42 |
ENST00000377470.3
ENST00000447830.2 |
SPATC1
|
spermatogenesis and centriole associated 1 |
| chr2_-_61389240 | 1.41 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr22_-_20138302 | 1.40 |
ENST00000540078.1
ENST00000439765.2 |
AC006547.14
|
uncharacterized protein LOC388849 |
| chr1_+_45212051 | 1.39 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr18_+_61143994 | 1.38 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr5_+_118965244 | 1.38 |
ENST00000515256.1
ENST00000509264.1 |
FAM170A
|
family with sequence similarity 170, member A |
| chr19_+_49866331 | 1.37 |
ENST00000597873.1
|
DKKL1
|
dickkopf-like 1 |
| chr22_+_23077065 | 1.36 |
ENST00000390310.2
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr9_+_139874683 | 1.35 |
ENST00000444903.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr9_-_88874519 | 1.35 |
ENST00000376001.3
ENST00000339137.3 |
C9orf153
|
chromosome 9 open reading frame 153 |
| chrX_+_23928500 | 1.34 |
ENST00000435707.1
|
CXorf58
|
chromosome X open reading frame 58 |
| chr7_+_23749767 | 1.32 |
ENST00000355870.3
|
STK31
|
serine/threonine kinase 31 |
| chr22_+_22453093 | 1.32 |
ENST00000390283.2
|
IGLV8-61
|
immunoglobulin lambda variable 8-61 |
| chr1_+_241815577 | 1.32 |
ENST00000366552.2
ENST00000437684.2 |
WDR64
|
WD repeat domain 64 |
| chrX_-_152989798 | 1.31 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr22_-_17073700 | 1.30 |
ENST00000359963.3
|
CCT8L2
|
chaperonin containing TCP1, subunit 8 (theta)-like 2 |
| chr17_+_48046538 | 1.30 |
ENST00000240306.3
|
DLX4
|
distal-less homeobox 4 |
| chrX_+_13587712 | 1.27 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr5_+_170288856 | 1.25 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr8_-_99955042 | 1.24 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr7_+_23749894 | 1.24 |
ENST00000433467.2
|
STK31
|
serine/threonine kinase 31 |
| chr12_+_48876275 | 1.24 |
ENST00000314014.2
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr2_+_220492116 | 1.24 |
ENST00000373760.2
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr22_-_29138386 | 1.23 |
ENST00000544772.1
|
CHEK2
|
checkpoint kinase 2 |
| chr2_-_231860596 | 1.22 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr14_-_89021077 | 1.21 |
ENST00000556564.1
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr14_-_106174960 | 1.21 |
ENST00000390547.2
|
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr14_+_27342334 | 1.18 |
ENST00000548170.1
ENST00000552926.1 |
RP11-384J4.1
|
RP11-384J4.1 |
| chr3_+_45067659 | 1.18 |
ENST00000296130.4
|
CLEC3B
|
C-type lectin domain family 3, member B |
| chr1_+_55107449 | 1.18 |
ENST00000421030.2
ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7
|
maestro heat-like repeat family member 7 |
| chr2_+_197577841 | 1.16 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr22_+_23134974 | 1.11 |
ENST00000390314.2
|
IGLV2-11
|
immunoglobulin lambda variable 2-11 |
| chr22_+_22569155 | 1.10 |
ENST00000390287.2
|
IGLV10-54
|
immunoglobulin lambda variable 10-54 |
| chr12_+_53817633 | 1.09 |
ENST00000257863.4
ENST00000550311.1 ENST00000379791.3 |
AMHR2
|
anti-Mullerian hormone receptor, type II |
| chr8_+_98881268 | 1.09 |
ENST00000254898.5
ENST00000524308.1 ENST00000522025.2 |
MATN2
|
matrilin 2 |
| chr1_-_32827682 | 1.08 |
ENST00000432622.1
|
FAM229A
|
family with sequence similarity 229, member A |
| chr15_-_31393910 | 1.07 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr3_+_14989186 | 1.06 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr22_+_22712087 | 1.06 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr4_+_20255123 | 1.05 |
ENST00000504154.1
ENST00000273739.5 |
SLIT2
|
slit homolog 2 (Drosophila) |
| chr12_+_52626898 | 1.03 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr18_-_19748379 | 1.03 |
ENST00000579431.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr12_+_102513950 | 1.03 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chr13_+_113030658 | 1.03 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr12_-_78753496 | 1.01 |
ENST00000548512.1
|
RP11-38F22.1
|
RP11-38F22.1 |
| chr22_+_23243156 | 1.01 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr3_-_51813009 | 1.00 |
ENST00000398780.3
|
IQCF6
|
IQ motif containing F6 |
| chr1_-_7913089 | 1.00 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr22_+_23040274 | 0.99 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr2_+_87565634 | 0.99 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr4_+_48343339 | 0.98 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr3_-_49726104 | 0.96 |
ENST00000383728.3
ENST00000545762.1 |
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr12_+_106994905 | 0.95 |
ENST00000357881.4
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr15_-_83316254 | 0.94 |
ENST00000567678.1
ENST00000450751.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr7_+_23749854 | 0.94 |
ENST00000456014.2
|
STK31
|
serine/threonine kinase 31 |
| chr1_+_196621156 | 0.91 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr2_+_179316163 | 0.90 |
ENST00000409117.3
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr18_-_19748331 | 0.89 |
ENST00000584201.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chrX_+_101380642 | 0.89 |
ENST00000372780.1
ENST00000329035.2 |
TCEAL2
|
transcription elongation factor A (SII)-like 2 |
| chr12_+_27849378 | 0.88 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr18_+_61144160 | 0.87 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr6_+_21593972 | 0.87 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr11_-_3240043 | 0.86 |
ENST00000332314.3
|
MRGPRG
|
MAS-related GPR, member G |
| chr1_+_55107489 | 0.86 |
ENST00000395690.2
|
MROH7
|
maestro heat-like repeat family member 7 |
| chr22_+_22676808 | 0.84 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr7_+_23749822 | 0.84 |
ENST00000422637.1
|
STK31
|
serine/threonine kinase 31 |
| chr1_-_103574024 | 0.83 |
ENST00000512756.1
ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1
|
collagen, type XI, alpha 1 |
| chr4_+_93225550 | 0.83 |
ENST00000282020.4
|
GRID2
|
glutamate receptor, ionotropic, delta 2 |
| chr17_+_37894179 | 0.82 |
ENST00000577695.1
ENST00000309156.4 ENST00000309185.3 |
GRB7
|
growth factor receptor-bound protein 7 |
| chr14_-_25078864 | 0.82 |
ENST00000216338.4
ENST00000557220.2 ENST00000382548.4 |
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX) |
| chr11_+_1860200 | 0.81 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr2_-_90538397 | 0.80 |
ENST00000443397.3
|
RP11-685N3.1
|
Uncharacterized protein |
| chr14_+_77292715 | 0.79 |
ENST00000393774.3
ENST00000555189.1 ENST00000450042.2 |
C14orf166B
|
chromosome 14 open reading frame 166B |
| chr17_-_58499698 | 0.79 |
ENST00000590297.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr5_-_145483932 | 0.79 |
ENST00000311450.4
|
PLAC8L1
|
PLAC8-like 1 |
| chr16_+_85942594 | 0.78 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr1_-_149900122 | 0.77 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr14_+_88852059 | 0.77 |
ENST00000045347.7
|
SPATA7
|
spermatogenesis associated 7 |
| chr7_+_128312346 | 0.75 |
ENST00000480462.1
ENST00000378704.3 ENST00000477515.1 |
FAM71F2
|
family with sequence similarity 71, member F2 |
| chr16_+_67700673 | 0.75 |
ENST00000403458.4
ENST00000602365.1 |
C16orf86
|
chromosome 16 open reading frame 86 |
| chr19_+_41117770 | 0.75 |
ENST00000601032.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr1_+_152881014 | 0.74 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr14_-_101351184 | 0.73 |
ENST00000534062.1
|
RTL1
|
retrotransposon-like 1 |
| chr3_+_172361483 | 0.73 |
ENST00000598405.1
|
AC007919.2
|
HCG1787166; PRO1163; Uncharacterized protein |
| chr7_+_76751926 | 0.73 |
ENST00000285871.4
ENST00000431197.1 |
CCDC146
|
coiled-coil domain containing 146 |
| chr10_+_76586348 | 0.73 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr10_+_77056181 | 0.73 |
ENST00000526759.1
ENST00000533822.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chr1_+_200842083 | 0.72 |
ENST00000304244.2
|
GPR25
|
G protein-coupled receptor 25 |
| chr3_-_79068594 | 0.71 |
ENST00000436010.2
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr6_-_31782813 | 0.70 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr12_-_52995179 | 0.70 |
ENST00000549979.1
|
KRT72
|
keratin 72 |
| chr1_-_38230738 | 0.70 |
ENST00000427468.2
ENST00000373048.4 ENST00000319637.6 |
EPHA10
|
EPH receptor A10 |
| chr12_-_133812681 | 0.69 |
ENST00000545940.1
|
ANHX
|
anomalous homeobox |
| chr11_+_64008525 | 0.69 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr1_-_242612779 | 0.68 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr15_-_83316087 | 0.68 |
ENST00000568757.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr8_-_144651024 | 0.68 |
ENST00000524906.1
ENST00000532862.1 ENST00000534459.1 |
MROH6
|
maestro heat-like repeat family member 6 |
| chr12_+_41221794 | 0.68 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr22_-_20255212 | 0.67 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr19_+_45281118 | 0.67 |
ENST00000270279.3
ENST00000341505.4 |
CBLC
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr19_+_32836499 | 0.66 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr1_+_196621002 | 0.66 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr2_+_39117010 | 0.66 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr3_+_138153451 | 0.64 |
ENST00000389567.4
ENST00000289135.4 |
ESYT3
|
extended synaptotagmin-like protein 3 |
| chr19_+_39989535 | 0.64 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr17_+_37894570 | 0.61 |
ENST00000394211.3
|
GRB7
|
growth factor receptor-bound protein 7 |
| chr9_+_133259836 | 0.60 |
ENST00000455439.2
|
HMCN2
|
hemicentin 2 |
| chr17_-_15496722 | 0.59 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr2_-_49381646 | 0.58 |
ENST00000346173.3
ENST00000406846.2 |
FSHR
|
follicle stimulating hormone receptor |
| chr12_-_95510743 | 0.57 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr16_+_28875126 | 0.57 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr12_-_50790267 | 0.56 |
ENST00000327337.5
ENST00000543111.1 |
FAM186A
|
family with sequence similarity 186, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of THRA_RXRB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 18.9 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 1.2 | 7.2 | GO:0042335 | cuticle development(GO:0042335) |
| 1.2 | 3.6 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 1.0 | 4.2 | GO:0060032 | notochord regression(GO:0060032) |
| 0.7 | 2.7 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.6 | 2.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.5 | 1.6 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.5 | 1.5 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.4 | 3.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.4 | 6.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.4 | 2.9 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.4 | 1.8 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.4 | 1.8 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.3 | 2.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 1.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.3 | 3.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 1.1 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.2 | 2.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 0.9 | GO:0038194 | thyroid-stimulating hormone signaling pathway(GO:0038194) cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.2 | 0.9 | GO:0035905 | N-terminal peptidyl-lysine acetylation(GO:0018076) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.2 | 0.8 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.2 | 1.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 3.5 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 3.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 1.7 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 1.3 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 1.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.2 | 2.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 0.5 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.2 | 0.2 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.2 | 1.1 | GO:0014826 | cellular magnesium ion homeostasis(GO:0010961) vein smooth muscle contraction(GO:0014826) |
| 0.2 | 0.5 | GO:1900211 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.1 | 0.7 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.8 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 1.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 2.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.5 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.1 | 0.9 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 3.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 2.6 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 2.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.8 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.5 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.1 | 13.2 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 1.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.2 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 1.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 2.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 1.6 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.4 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.2 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.1 | 0.3 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.6 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 1.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 12.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.3 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.2 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) ganglion morphogenesis(GO:0061552) blood vessel endothelial cell fate specification(GO:0097101) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.2 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.7 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 2.2 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 4.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 1.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 3.2 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.5 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 1.4 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.8 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 5.1 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 3.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 2.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0036376 | sodium ion export from cell(GO:0036376) regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 0.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 1.6 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 | 2.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 1.1 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.3 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.9 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.0 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.8 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 1.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.6 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 1.0 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 0.7 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0071745 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 1.0 | 2.9 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.3 | 18.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.3 | 1.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.3 | 3.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 1.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.2 | 1.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 7.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 1.2 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 2.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 2.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 10.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 4.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 2.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.3 | GO:1990234 | transferase complex(GO:1990234) |
| 0.1 | 5.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 2.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 1.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.7 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.3 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 2.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 5.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 2.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 3.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 9.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.0 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 5.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.6 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0004040 | amidase activity(GO:0004040) |
| 0.4 | 2.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.4 | 6.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.4 | 5.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 1.1 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.3 | 1.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.3 | 1.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.3 | 1.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.3 | 2.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 0.7 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.2 | 1.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.6 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.2 | 2.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.5 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.1 | 1.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.9 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.1 | 0.5 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.1 | 2.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 7.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 3.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 3.5 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 18.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 3.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 2.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.2 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 1.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 1.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 3.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 1.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 1.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 5.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.0 | 2.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 6.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.0 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.0 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 5.8 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 3.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 1.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 2.7 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 5.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 3.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 3.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 5.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 4.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 3.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 4.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 2.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 4.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 2.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |