Project
Illumina Body Map 2
Navigation
Downloads
Results for THRB
Z-value: 0.85
Transcription factors associated with THRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRB
|
ENSG00000151090.13 | thyroid hormone receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| THRB | hg19_v2_chr3_-_24536222_24536249 | -0.82 | 6.5e-09 | Click! |
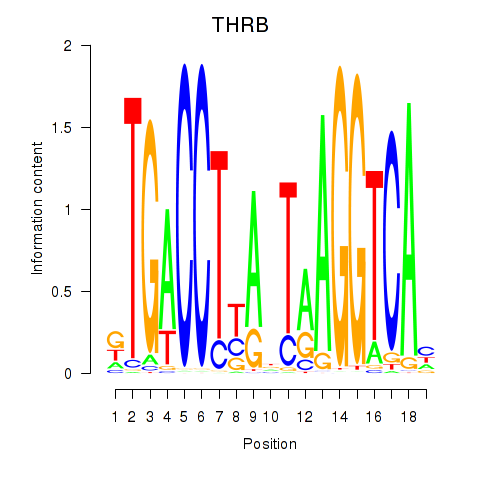
Activity profile of THRB motif
Sorted Z-values of THRB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_22180536 | 4.52 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr19_+_55085248 | 3.46 |
ENST00000391738.3
ENST00000251376.3 ENST00000391737.1 ENST00000396321.2 ENST00000418536.2 ENST00000448689.1 |
LILRA2
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2 leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
| chr17_-_73840415 | 3.42 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr20_+_37434329 | 3.24 |
ENST00000299824.1
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B |
| chr17_-_73840614 | 3.12 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr11_-_47400062 | 3.11 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr7_-_38394118 | 3.08 |
ENST00000390345.2
|
TRGV4
|
T cell receptor gamma variable 4 |
| chr11_-_47399942 | 3.00 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr11_-_47400032 | 2.85 |
ENST00000533968.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr11_-_47400078 | 2.71 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr17_-_73840774 | 2.65 |
ENST00000207549.4
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr17_-_73839792 | 2.35 |
ENST00000590762.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr6_+_42883727 | 2.19 |
ENST00000304672.1
ENST00000441198.1 ENST00000446507.1 |
PTCRA
|
pre T-cell antigen receptor alpha |
| chr8_-_131028782 | 2.16 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr8_+_28174649 | 2.01 |
ENST00000301908.3
|
PNOC
|
prepronociceptin |
| chr16_+_72088376 | 1.98 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr8_+_28174496 | 1.77 |
ENST00000518479.1
|
PNOC
|
prepronociceptin |
| chr2_-_73869508 | 1.46 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr9_-_139268068 | 1.41 |
ENST00000371734.3
ENST00000371732.5 ENST00000315908.7 |
CARD9
|
caspase recruitment domain family, member 9 |
| chr11_+_71934962 | 1.26 |
ENST00000543234.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr8_-_124037890 | 1.26 |
ENST00000519018.1
ENST00000523036.1 |
DERL1
|
derlin 1 |
| chr11_+_5617952 | 1.22 |
ENST00000354852.5
|
TRIM6-TRIM34
|
TRIM6-TRIM34 readthrough |
| chrX_-_1511617 | 1.07 |
ENST00000381401.5
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chrY_-_1461617 | 1.03 |
ENSTR0000381401.5
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr2_-_74692473 | 0.98 |
ENST00000535045.1
ENST00000409065.1 ENST00000414701.1 ENST00000448666.1 ENST00000233616.4 ENST00000452063.2 |
MOGS
|
mannosyl-oligosaccharide glucosidase |
| chr6_+_77484663 | 0.98 |
ENST00000607287.1
|
RP11-354K4.2
|
RP11-354K4.2 |
| chr12_+_54378849 | 0.92 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr11_+_73498973 | 0.84 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr13_+_27844464 | 0.78 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr1_-_154164534 | 0.72 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr1_+_161190160 | 0.71 |
ENST00000594609.1
|
AL590714.1
|
Uncharacterized protein |
| chr14_+_75761099 | 0.71 |
ENST00000561000.1
ENST00000558575.1 |
RP11-293M10.5
|
RP11-293M10.5 |
| chr15_-_101084446 | 0.68 |
ENST00000538112.2
ENST00000559639.1 ENST00000558884.2 |
CERS3
|
ceramide synthase 3 |
| chr10_+_60145155 | 0.67 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr11_+_73498898 | 0.66 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr17_+_1646130 | 0.64 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr16_-_69368774 | 0.62 |
ENST00000562949.1
|
RP11-343C2.12
|
Conserved oligomeric Golgi complex subunit 8 |
| chr12_+_54378923 | 0.50 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr21_-_37914898 | 0.49 |
ENST00000399136.1
|
CLDN14
|
claudin 14 |
| chr1_+_77997785 | 0.49 |
ENST00000478255.1
|
AK5
|
adenylate kinase 5 |
| chr11_-_18747727 | 0.38 |
ENST00000513874.1
|
IGSF22
|
immunoglobulin superfamily, member 22 |
| chr19_-_52643157 | 0.38 |
ENST00000597013.1
ENST00000600228.1 ENST00000596290.1 |
ZNF616
|
zinc finger protein 616 |
| chr3_+_172361483 | 0.32 |
ENST00000598405.1
|
AC007919.2
|
HCG1787166; PRO1163; Uncharacterized protein |
| chr11_-_64014379 | 0.30 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_-_207224307 | 0.28 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr22_+_19938419 | 0.25 |
ENST00000412786.1
|
COMT
|
catechol-O-methyltransferase |
| chr18_-_47808050 | 0.24 |
ENST00000590208.1
|
MBD1
|
methyl-CpG binding domain protein 1 |
| chr18_-_47808120 | 0.24 |
ENST00000587605.1
ENST00000591416.1 ENST00000382948.5 ENST00000269471.5 ENST00000269468.5 ENST00000347968.3 ENST00000436910.1 ENST00000349085.2 |
MBD1
|
methyl-CpG binding domain protein 1 |
| chr15_-_35838348 | 0.19 |
ENST00000561411.1
ENST00000256538.4 ENST00000440392.2 |
DPH6
|
diphthamine biosynthesis 6 |
| chr8_+_19674651 | 0.18 |
ENST00000397977.3
|
INTS10
|
integrator complex subunit 10 |
| chr5_+_131746575 | 0.18 |
ENST00000337752.2
ENST00000378947.3 ENST00000407797.1 |
C5orf56
|
chromosome 5 open reading frame 56 |
| chr6_+_151042224 | 0.17 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr20_+_388679 | 0.11 |
ENST00000356286.5
ENST00000475269.1 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr12_-_21810765 | 0.06 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr20_+_388791 | 0.05 |
ENST00000441733.1
ENST00000353660.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr13_-_24007815 | 0.03 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr18_-_47807829 | 0.02 |
ENST00000585672.1
ENST00000457839.2 ENST00000353909.3 ENST00000339998.6 ENST00000398493.1 |
MBD1
|
methyl-CpG binding domain protein 1 |
| chr8_-_100905850 | 0.01 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr2_-_9563216 | 0.01 |
ENST00000467606.1
ENST00000494563.1 ENST00000460001.1 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr2_-_9563469 | 0.00 |
ENST00000484735.1
ENST00000456913.2 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr15_-_101084547 | 0.00 |
ENST00000394113.1
|
CERS3
|
ceramide synthase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of THRB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.5 | GO:0002432 | granuloma formation(GO:0002432) |
| 2.3 | 11.7 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 1.1 | 3.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.9 | 3.5 | GO:2001193 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.7 | 2.1 | GO:0052510 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.7 | 2.0 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.3 | 2.2 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.2 | 1.3 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.2 | 0.7 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 0.5 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.1 | 0.6 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 1.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.4 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.3 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 3.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.5 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 4.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.8 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.5 | 2.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 1.3 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.1 | 2.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 1.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 11.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 3.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 2.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 11.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.9 | 3.5 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.5 | 3.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.5 | 2.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.3 | 2.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 2.2 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.2 | 1.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.7 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.1 | 1.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 0.3 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 1.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.0 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.5 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 11.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 3.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 2.2 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 2.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 3.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 3.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |