Project
Illumina Body Map 2
Navigation
Downloads
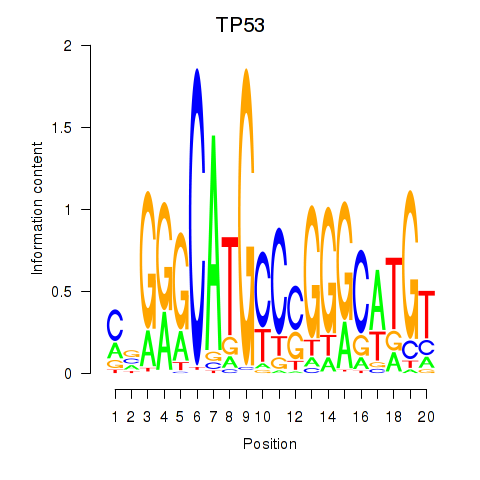
Results for TP53
Z-value: 1.39
Transcription factors associated with TP53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP53
|
ENSG00000141510.11 | tumor protein p53 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP53 | hg19_v2_chr17_-_7590745_7590856 | 0.39 | 2.6e-02 | Click! |
Activity profile of TP53 motif
Sorted Z-values of TP53 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_89417335 | 4.57 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr8_+_22960426 | 4.13 |
ENST00000540813.1
|
TNFRSF10C
|
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain |
| chr14_-_106692191 | 3.92 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr19_+_827823 | 2.75 |
ENST00000233997.2
|
AZU1
|
azurocidin 1 |
| chr19_-_6591113 | 2.68 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr6_+_36098262 | 2.57 |
ENST00000373761.6
ENST00000373766.5 |
MAPK13
|
mitogen-activated protein kinase 13 |
| chr11_+_289110 | 2.51 |
ENST00000409548.2
|
ATHL1
|
ATH1, acid trehalase-like 1 (yeast) |
| chr11_+_289155 | 2.46 |
ENST00000409655.1
|
ATHL1
|
ATH1, acid trehalase-like 1 (yeast) |
| chr8_-_23021533 | 2.45 |
ENST00000312584.3
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
| chr11_-_47400078 | 2.30 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr11_-_47400032 | 2.26 |
ENST00000533968.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr11_-_47399942 | 2.22 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr8_-_23082580 | 2.20 |
ENST00000221132.3
|
TNFRSF10A
|
tumor necrosis factor receptor superfamily, member 10a |
| chr11_-_47400062 | 2.16 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr6_-_30712313 | 2.11 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr11_-_44972390 | 2.08 |
ENST00000395648.3
ENST00000531928.2 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr1_-_54483842 | 1.98 |
ENST00000371362.3
ENST00000420619.1 |
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1 |
| chr18_+_57567180 | 1.97 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr19_+_3136115 | 1.96 |
ENST00000262958.3
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
| chr1_-_54483803 | 1.83 |
ENST00000371360.1
ENST00000545928.1 |
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1 |
| chr11_-_44972476 | 1.76 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr11_+_67171391 | 1.75 |
ENST00000312390.5
|
TBC1D10C
|
TBC1 domain family, member 10C |
| chr1_-_160832490 | 1.75 |
ENST00000322302.7
ENST00000368033.3 |
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr17_+_76210267 | 1.70 |
ENST00000301633.4
ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr11_+_67171358 | 1.70 |
ENST00000526387.1
|
TBC1D10C
|
TBC1 domain family, member 10C |
| chr1_-_209825674 | 1.69 |
ENST00000367030.3
ENST00000356082.4 |
LAMB3
|
laminin, beta 3 |
| chr3_-_52479043 | 1.69 |
ENST00000231721.2
ENST00000475739.1 |
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chr19_+_49458107 | 1.67 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr3_+_122785895 | 1.64 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr1_-_160832642 | 1.61 |
ENST00000368034.4
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr20_+_33759854 | 1.60 |
ENST00000216968.4
|
PROCR
|
protein C receptor, endothelial |
| chr1_-_38412683 | 1.58 |
ENST00000373024.3
ENST00000373023.2 |
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa |
| chr20_-_1373726 | 1.55 |
ENST00000400137.4
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr1_-_153588334 | 1.52 |
ENST00000476873.1
|
S100A14
|
S100 calcium binding protein A14 |
| chr17_-_61523535 | 1.52 |
ENST00000584031.1
ENST00000392976.1 |
CYB561
|
cytochrome b561 |
| chr10_-_105212059 | 1.52 |
ENST00000260743.5
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr20_-_1373606 | 1.51 |
ENST00000381715.1
ENST00000439640.2 ENST00000381719.3 |
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chrX_-_153583257 | 1.48 |
ENST00000438732.1
|
FLNA
|
filamin A, alpha |
| chr12_-_53207842 | 1.48 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr10_+_47746929 | 1.47 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr1_+_10057274 | 1.46 |
ENST00000294435.7
|
RBP7
|
retinol binding protein 7, cellular |
| chr2_-_157198860 | 1.45 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr11_+_5712234 | 1.41 |
ENST00000414641.1
|
TRIM22
|
tripartite motif containing 22 |
| chr6_+_36097992 | 1.41 |
ENST00000211287.4
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chrX_+_99899180 | 1.40 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr8_-_22926526 | 1.39 |
ENST00000347739.3
ENST00000542226.1 |
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b |
| chr10_+_48255253 | 1.38 |
ENST00000357718.4
ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8
AL591684.1
|
annexin A8 Protein LOC100996760 |
| chr17_-_61523622 | 1.37 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr19_+_42364460 | 1.35 |
ENST00000593863.1
|
RPS19
|
ribosomal protein S19 |
| chr8_-_10588010 | 1.34 |
ENST00000304501.1
|
SOX7
|
SRY (sex determining region Y)-box 7 |
| chr17_+_76210367 | 1.32 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr19_-_6481776 | 1.29 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr5_-_131826457 | 1.28 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr3_+_184279566 | 1.27 |
ENST00000330394.2
|
EPHB3
|
EPH receptor B3 |
| chr20_-_1373682 | 1.27 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr10_+_90750493 | 1.26 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr11_+_706595 | 1.26 |
ENST00000531348.1
ENST00000530636.1 |
EPS8L2
|
EPS8-like 2 |
| chr7_+_100770328 | 1.25 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr1_-_160832670 | 1.25 |
ENST00000368032.2
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr11_-_44972299 | 1.24 |
ENST00000528473.1
|
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr10_-_105212141 | 1.24 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr11_+_36397528 | 1.24 |
ENST00000311599.5
ENST00000378867.3 |
PRR5L
|
proline rich 5 like |
| chr8_-_22926623 | 1.21 |
ENST00000276431.4
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b |
| chr11_-_65626797 | 1.20 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr19_+_42364313 | 1.20 |
ENST00000601492.1
ENST00000600467.1 ENST00000221975.2 |
RPS19
|
ribosomal protein S19 |
| chr1_-_156051789 | 1.19 |
ENST00000532414.2
|
MEX3A
|
mex-3 RNA binding family member A |
| chr1_+_28206150 | 1.19 |
ENST00000456990.1
|
THEMIS2
|
thymocyte selection associated family member 2 |
| chr1_-_150738261 | 1.19 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr22_-_18257178 | 1.18 |
ENST00000342111.5
|
BID
|
BH3 interacting domain death agonist |
| chr11_-_65626753 | 1.17 |
ENST00000526975.1
ENST00000531413.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr1_+_25599018 | 1.17 |
ENST00000417538.2
ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD
|
Rh blood group, D antigen |
| chr2_+_208104497 | 1.16 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr18_-_28622774 | 1.15 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr12_+_22778291 | 1.14 |
ENST00000545979.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr14_-_36990354 | 1.13 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr6_+_36098363 | 1.12 |
ENST00000373759.1
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chr17_+_74372662 | 1.11 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr5_+_121647386 | 1.10 |
ENST00000542191.1
ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr1_+_2477831 | 1.10 |
ENST00000606645.1
|
RP3-395M20.12
|
RP3-395M20.12 |
| chr18_-_28622699 | 1.10 |
ENST00000360428.4
|
DSC3
|
desmocollin 3 |
| chr11_+_124761200 | 1.09 |
ENST00000524433.1
|
RP11-664I21.6
|
Uncharacterized protein |
| chr19_+_3585471 | 1.09 |
ENST00000322315.5
|
GIPC3
|
GIPC PDZ domain containing family, member 3 |
| chr7_-_72439997 | 1.09 |
ENST00000285805.3
|
TRIM74
|
tripartite motif containing 74 |
| chr4_+_153457404 | 1.09 |
ENST00000604157.1
ENST00000594836.1 |
MIR4453
|
microRNA 4453 |
| chr2_+_95691445 | 1.08 |
ENST00000353004.3
ENST00000354078.3 ENST00000349807.3 |
MAL
|
mal, T-cell differentiation protein |
| chr20_+_30946106 | 1.06 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr19_+_18492973 | 1.05 |
ENST00000595973.2
|
GDF15
|
growth differentiation factor 15 |
| chr11_-_73694346 | 1.05 |
ENST00000310473.3
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr22_+_37447771 | 1.03 |
ENST00000402077.3
ENST00000403888.3 ENST00000456470.1 |
KCTD17
|
potassium channel tetramerization domain containing 17 |
| chr9_-_130517309 | 1.03 |
ENST00000414380.1
|
SH2D3C
|
SH2 domain containing 3C |
| chr4_-_10118348 | 1.02 |
ENST00000502702.1
|
WDR1
|
WD repeat domain 1 |
| chr2_+_95691417 | 1.01 |
ENST00000309988.4
|
MAL
|
mal, T-cell differentiation protein |
| chr16_-_75467274 | 1.01 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr11_-_44972418 | 1.01 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr15_+_100347228 | 1.00 |
ENST00000559714.1
ENST00000560059.1 |
CTD-2054N24.2
|
Uncharacterized protein |
| chr19_+_42746927 | 0.97 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr14_-_75078725 | 0.97 |
ENST00000556690.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr4_-_10118573 | 0.96 |
ENST00000382452.2
ENST00000382451.2 |
WDR1
|
WD repeat domain 1 |
| chr10_-_47173994 | 0.95 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr7_+_75024903 | 0.95 |
ENST00000323819.3
ENST00000430211.1 |
TRIM73
|
tripartite motif containing 73 |
| chr16_-_14788526 | 0.95 |
ENST00000438167.3
|
PLA2G10
|
phospholipase A2, group X |
| chr7_+_116165754 | 0.94 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr1_+_17634689 | 0.94 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr1_-_54518865 | 0.92 |
ENST00000371337.3
|
TMEM59
|
transmembrane protein 59 |
| chr11_+_61717535 | 0.91 |
ENST00000534553.1
ENST00000301774.9 |
BEST1
|
bestrophin 1 |
| chr11_+_827553 | 0.91 |
ENST00000528542.2
ENST00000450448.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr2_+_11295498 | 0.90 |
ENST00000295083.3
ENST00000441908.2 |
PQLC3
|
PQ loop repeat containing 3 |
| chr3_+_14166440 | 0.88 |
ENST00000306077.4
|
TMEM43
|
transmembrane protein 43 |
| chr17_+_25621102 | 0.88 |
ENST00000581440.1
ENST00000262394.2 ENST00000583742.1 ENST00000579733.1 ENST00000583193.1 ENST00000581185.1 ENST00000427287.2 ENST00000348811.2 |
WSB1
|
WD repeat and SOCS box containing 1 |
| chr5_+_121647877 | 0.88 |
ENST00000514497.2
ENST00000261367.7 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr4_-_10117949 | 0.87 |
ENST00000508079.1
|
WDR1
|
WD repeat domain 1 |
| chr19_+_35861831 | 0.87 |
ENST00000454971.1
|
GPR42
|
G protein-coupled receptor 42 (gene/pseudogene) |
| chr1_-_154941350 | 0.85 |
ENST00000444179.1
ENST00000414115.1 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr19_-_42721819 | 0.84 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr1_+_3607228 | 0.84 |
ENST00000378285.1
ENST00000378280.1 ENST00000378288.4 |
TP73
|
tumor protein p73 |
| chr14_-_75079026 | 0.84 |
ENST00000261978.4
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr1_-_113249734 | 0.83 |
ENST00000484054.3
ENST00000369636.2 ENST00000369637.1 ENST00000285735.2 ENST00000369638.2 |
RHOC
|
ras homolog family member C |
| chr12_-_14721283 | 0.82 |
ENST00000240617.5
|
PLBD1
|
phospholipase B domain containing 1 |
| chr4_-_129209221 | 0.82 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr19_+_56652556 | 0.81 |
ENST00000337080.3
|
ZNF444
|
zinc finger protein 444 |
| chr2_+_11295624 | 0.81 |
ENST00000402361.1
ENST00000428481.1 |
PQLC3
|
PQ loop repeat containing 3 |
| chr22_-_39715600 | 0.81 |
ENST00000427905.1
ENST00000402527.1 ENST00000216146.4 |
RPL3
|
ribosomal protein L3 |
| chr22_+_42949925 | 0.81 |
ENST00000327678.5
ENST00000340239.4 ENST00000407614.4 ENST00000335879.5 |
SERHL2
|
serine hydrolase-like 2 |
| chr12_-_113772835 | 0.81 |
ENST00000552014.1
ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr1_+_11796126 | 0.81 |
ENST00000376637.3
|
AGTRAP
|
angiotensin II receptor-associated protein |
| chr1_-_113249678 | 0.80 |
ENST00000369633.2
ENST00000425265.2 ENST00000369632.2 ENST00000436685.2 |
RHOC
|
ras homolog family member C |
| chr16_-_15149917 | 0.80 |
ENST00000287706.3
|
NTAN1
|
N-terminal asparagine amidase |
| chrX_-_65858865 | 0.80 |
ENST00000374719.3
ENST00000450752.1 ENST00000451436.2 |
EDA2R
|
ectodysplasin A2 receptor |
| chr13_+_31480328 | 0.80 |
ENST00000380482.4
|
MEDAG
|
mesenteric estrogen-dependent adipogenesis |
| chr19_-_51530916 | 0.79 |
ENST00000594768.1
|
KLK11
|
kallikrein-related peptidase 11 |
| chr19_+_55999771 | 0.79 |
ENST00000594321.1
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains) |
| chr2_+_177053307 | 0.78 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr17_-_74528128 | 0.78 |
ENST00000590175.1
|
CYGB
|
cytoglobin |
| chr5_+_27472371 | 0.77 |
ENST00000512067.1
ENST00000510165.1 ENST00000505775.1 ENST00000514844.1 ENST00000503107.1 |
LINC01021
|
long intergenic non-protein coding RNA 1021 |
| chr16_-_75467318 | 0.77 |
ENST00000283882.3
|
CFDP1
|
craniofacial development protein 1 |
| chr19_+_35849723 | 0.77 |
ENST00000594310.1
|
FFAR3
|
free fatty acid receptor 3 |
| chr9_-_135996537 | 0.76 |
ENST00000372050.3
ENST00000372047.3 |
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr22_-_50708781 | 0.76 |
ENST00000449719.2
ENST00000330651.6 |
MAPK11
|
mitogen-activated protein kinase 11 |
| chr1_-_3528034 | 0.76 |
ENST00000356575.4
|
MEGF6
|
multiple EGF-like-domains 6 |
| chr19_-_6481759 | 0.76 |
ENST00000588421.1
|
DENND1C
|
DENN/MADD domain containing 1C |
| chr11_+_827248 | 0.76 |
ENST00000527089.1
ENST00000530183.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr3_+_52529346 | 0.76 |
ENST00000321725.6
|
STAB1
|
stabilin 1 |
| chr19_+_35862192 | 0.75 |
ENST00000597214.1
|
GPR42
|
G protein-coupled receptor 42 (gene/pseudogene) |
| chr19_+_55999916 | 0.74 |
ENST00000587166.1
ENST00000389623.6 |
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains) |
| chr17_-_9929581 | 0.74 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr19_-_28284793 | 0.73 |
ENST00000590523.1
|
LINC00662
|
long intergenic non-protein coding RNA 662 |
| chr12_+_69186125 | 0.73 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr3_-_58196688 | 0.73 |
ENST00000486455.1
|
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr19_-_47735918 | 0.72 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr1_-_155990580 | 0.72 |
ENST00000531917.1
ENST00000480567.1 ENST00000526212.1 ENST00000529008.1 ENST00000496742.1 ENST00000295702.4 |
SSR2
|
signal sequence receptor, beta (translocon-associated protein beta) |
| chrX_+_192989 | 0.71 |
ENST00000399012.1
ENST00000430923.2 |
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr20_+_43595115 | 0.70 |
ENST00000372806.3
ENST00000396731.4 ENST00000372801.1 ENST00000499879.2 |
STK4
|
serine/threonine kinase 4 |
| chr5_+_121647764 | 0.70 |
ENST00000261368.8
ENST00000379533.2 ENST00000379536.2 ENST00000379538.3 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr1_-_45805752 | 0.70 |
ENST00000354383.6
ENST00000355498.2 ENST00000372100.5 ENST00000531105.1 |
MUTYH
|
mutY homolog |
| chr1_+_25598989 | 0.69 |
ENST00000454452.2
|
RHD
|
Rh blood group, D antigen |
| chr4_-_10118469 | 0.69 |
ENST00000499869.2
|
WDR1
|
WD repeat domain 1 |
| chr19_-_52150053 | 0.68 |
ENST00000599649.1
ENST00000429354.3 ENST00000360844.6 ENST00000222107.4 |
SIGLEC5
SIGLEC14
SIGLEC5
|
SIGLEC5 sialic acid binding Ig-like lectin 14 sialic acid binding Ig-like lectin 5 |
| chr16_-_31106048 | 0.68 |
ENST00000300851.6
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr2_+_47168313 | 0.68 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr16_+_30207122 | 0.68 |
ENST00000395137.2
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr11_+_64126671 | 0.67 |
ENST00000530504.1
|
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
| chr1_+_27153173 | 0.66 |
ENST00000374142.4
|
ZDHHC18
|
zinc finger, DHHC-type containing 18 |
| chr16_-_31106211 | 0.66 |
ENST00000532364.1
ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1
VKORC1
|
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr11_+_65222698 | 0.66 |
ENST00000309775.7
|
AP000769.1
|
Uncharacterized protein |
| chr10_-_126847276 | 0.66 |
ENST00000531469.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr19_+_56652686 | 0.66 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr19_-_39926268 | 0.65 |
ENST00000599705.1
|
RPS16
|
ribosomal protein S16 |
| chr10_-_73611046 | 0.65 |
ENST00000394934.1
ENST00000394936.3 |
PSAP
|
prosaposin |
| chr17_+_14204389 | 0.65 |
ENST00000360954.2
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
| chr1_+_114447763 | 0.64 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr15_+_42867857 | 0.64 |
ENST00000290607.7
|
STARD9
|
StAR-related lipid transfer (START) domain containing 9 |
| chr6_+_116692102 | 0.64 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr14_-_36990061 | 0.63 |
ENST00000546983.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr15_-_42386752 | 0.63 |
ENST00000290472.3
|
PLA2G4D
|
phospholipase A2, group IVD (cytosolic) |
| chr22_+_21336267 | 0.62 |
ENST00000215739.8
|
LZTR1
|
leucine-zipper-like transcription regulator 1 |
| chr16_+_29467780 | 0.62 |
ENST00000395400.3
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr15_-_64455404 | 0.61 |
ENST00000300026.3
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B) |
| chr3_-_149510553 | 0.61 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr9_-_116037840 | 0.61 |
ENST00000374206.3
|
CDC26
|
cell division cycle 26 |
| chr19_-_17014408 | 0.61 |
ENST00000594249.1
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr2_+_109745655 | 0.59 |
ENST00000418513.1
|
SH3RF3
|
SH3 domain containing ring finger 3 |
| chr5_+_121647924 | 0.58 |
ENST00000414317.2
|
SNCAIP
|
synuclein, alpha interacting protein |
| chr11_+_64126614 | 0.57 |
ENST00000528057.1
ENST00000334205.4 ENST00000294261.4 |
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
| chr14_-_36789783 | 0.57 |
ENST00000605579.1
ENST00000604336.1 ENST00000359527.7 ENST00000603139.1 ENST00000318473.7 |
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr14_-_103989033 | 0.56 |
ENST00000553878.1
ENST00000557530.1 |
CKB
|
creatine kinase, brain |
| chr1_+_11796177 | 0.56 |
ENST00000400895.2
ENST00000376629.4 ENST00000376627.2 ENST00000314340.5 ENST00000452018.2 ENST00000510878.1 |
AGTRAP
|
angiotensin II receptor-associated protein |
| chr1_-_45805667 | 0.56 |
ENST00000488731.2
ENST00000435155.1 |
MUTYH
|
mutY homolog |
| chr1_-_45806053 | 0.55 |
ENST00000412971.1
ENST00000372098.3 ENST00000372110.3 ENST00000529984.1 ENST00000528332.2 ENST00000372115.3 ENST00000450313.1 |
MUTYH
|
mutY homolog |
| chr20_+_62289640 | 0.53 |
ENST00000508582.2
ENST00000360203.5 ENST00000356810.4 |
RTEL1
|
regulator of telomere elongation helicase 1 |
| chr8_-_69243711 | 0.52 |
ENST00000512294.3
|
RP11-664D7.4
|
HCG1787533; Uncharacterized protein |
| chr11_+_61717279 | 0.51 |
ENST00000378043.4
|
BEST1
|
bestrophin 1 |
| chr16_-_31105870 | 0.51 |
ENST00000394971.3
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr16_+_57406437 | 0.51 |
ENST00000564948.1
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr12_-_6451186 | 0.51 |
ENST00000540022.1
ENST00000536194.1 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr19_-_44259136 | 0.50 |
ENST00000270066.6
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr17_-_37308824 | 0.50 |
ENST00000415163.1
ENST00000441877.1 ENST00000444911.2 |
PLXDC1
|
plexin domain containing 1 |
| chr11_+_61717336 | 0.50 |
ENST00000378042.3
|
BEST1
|
bestrophin 1 |
| chr6_+_52285131 | 0.50 |
ENST00000433625.2
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr11_-_9286627 | 0.49 |
ENST00000530044.1
|
DENND5A
|
DENN/MADD domain containing 5A |
| chr11_-_68611721 | 0.49 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP53
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.9 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 1.2 | 4.6 | GO:0071613 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.9 | 2.7 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.9 | 3.5 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.6 | 2.6 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.6 | 1.3 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.6 | 1.7 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.5 | 1.5 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.4 | 1.3 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.4 | 1.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.4 | 1.5 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.4 | 1.8 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.4 | 1.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.3 | 3.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.3 | 1.4 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.3 | 2.4 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.3 | 4.3 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.3 | 0.9 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.3 | 1.6 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.3 | 0.8 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.3 | 0.5 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.3 | 1.5 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.3 | 1.0 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.2 | 6.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.2 | 1.8 | GO:0045007 | depurination(GO:0045007) |
| 0.2 | 2.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 1.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 | 0.6 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.2 | 0.6 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.2 | 0.8 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.2 | 0.9 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 1.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.2 | 0.9 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 4.8 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 1.8 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.2 | 1.9 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 1.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.8 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 0.8 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.4 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.2 | GO:1902731 | negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 3.0 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 1.9 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.8 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.6 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.4 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 0.6 | GO:0031860 | telomeric loop formation(GO:0031627) telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.6 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 1.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 1.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.7 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 1.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.4 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 3.3 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 0.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.4 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.1 | 0.3 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.2 | GO:0043132 | NAD transport(GO:0043132) |
| 0.1 | 1.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.2 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.1 | 0.4 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.1 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 1.5 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.4 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 8.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.5 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 0.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 1.9 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.5 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.3 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 1.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.2 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 5.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.7 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 1.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 1.0 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 1.7 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 1.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.5 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 5.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.2 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.7 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.8 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 1.3 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.5 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.4 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 1.0 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 3.4 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 2.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 1.4 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 1.3 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.6 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.9 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.6 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.6 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.5 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.3 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.7 | 3.5 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.4 | 3.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.4 | 1.5 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 1.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 1.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 1.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 1.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.9 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 3.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 3.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 2.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 2.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.8 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 1.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 1.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 3.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 1.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 3.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 3.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.7 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 4.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 10.2 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 1.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 4.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 1.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.4 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.7 | 2.9 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.7 | 8.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.5 | 1.8 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.4 | 1.9 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.4 | 2.6 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 1.0 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.3 | 0.9 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.3 | 1.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 4.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 1.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 6.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 1.4 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 0.8 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.2 | 0.9 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 0.5 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.2 | 0.5 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.2 | 1.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.4 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 1.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.6 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.6 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 2.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.6 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 0.7 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 4.6 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 1.8 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.3 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 2.6 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.1 | 0.7 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.8 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 3.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 2.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 2.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 3.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.7 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 2.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 1.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.3 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 1.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 1.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 2.3 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 1.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 1.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 1.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 2.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 4.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 1.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 3.7 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 6.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 3.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.1 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 4.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 3.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 11.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 3.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 9.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 3.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 3.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 3.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 4.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 5.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 2.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 4.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 3.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 3.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 3.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 4.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 5.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 3.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 2.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |